Blood pressure regulation by CD4+ lymphocytes expressing choline acetyltransferase
- PMID: 27617738
- PMCID: PMC5513182
- DOI: 10.1038/nbt.3663
Blood pressure regulation by CD4+ lymphocytes expressing choline acetyltransferase
Abstract
Blood pressure regulation is known to be maintained by a neuro-endocrine circuit, but whether immune cells contribute to blood pressure homeostasis has not been determined. We previously showed that CD4+ T lymphocytes that express choline acetyltransferase (ChAT), which catalyzes the synthesis of the vasorelaxant acetylcholine, relay neural signals. Here we show that these CD4+CD44hiCD62Llo T helper cells by gene expression are a distinct T-cell population defined by ChAT (CD4 TChAT). Mice lacking ChAT expression in CD4+ cells have elevated arterial blood pressure, compared to littermate controls. Jurkat T cells overexpressing ChAT (JTChAT) decreased blood pressure when infused into mice. Co-incubation of JTChAT and endothelial cells increased endothelial cell levels of phosphorylated endothelial nitric oxide synthase, and of nitrates and nitrites in conditioned media, indicating increased release of the potent vasorelaxant nitric oxide. The isolation and characterization of CD4 TChAT cells will enable analysis of the role of these cells in hypotension and hypertension, and may suggest novel therapeutic strategies by targeting cell-mediated vasorelaxation.
Conflict of interest statement
The authors have no competing financial interests to declare.
Figures
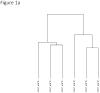





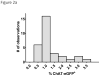



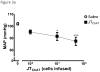


References
-
- SEVENTY-FOURTH ANNUAL MEETING OF THE British Medical Association. BMJ (Clinical research ed) 1906;2:1760–1816.
-
- Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. - PubMed
-
- Fleming I, Busse R. NO: the primary EDRF. J Mol Cell Cardiol. 1999;31:5–14. - PubMed
-
- Mulvany MJ, Aalkjaer C. Structure and function of small arteries. Physiol Rev. 1990;70:921–961. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

