The rules and impact of nonsense-mediated mRNA decay in human cancers
- PMID: 27618451
- PMCID: PMC5045715
- DOI: 10.1038/ng.3664
The rules and impact of nonsense-mediated mRNA decay in human cancers
Abstract
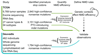
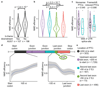
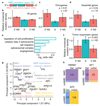
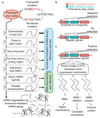
Premature termination codons (PTCs) cause a large proportion of inherited human genetic diseases. PTC-containing transcripts can be degraded by an mRNA surveillance pathway termed nonsense-mediated mRNA decay (NMD). However, the efficiency of NMD varies; it is inefficient when a PTC is located downstream of the last exon junction complex (EJC). We used matched exome and transcriptome data from 9,769 human tumors to systematically elucidate the rules of NMD targeting in human cells. An integrated model incorporating multiple rules beyond the canonical EJC model explains approximately three-fourths of the non-random variance in NMD efficiency across thousands of PTCs. We also show that dosage compensation may sometimes mask the effects of NMD. Applying the NMD model identifies signatures of both positive and negative selection on NMD-triggering mutations in human tumors and provides a classification for tumor-suppressor genes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–8. - PubMed
-
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–900. - PubMed
-
- Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood. 1994;83:2031–7. - PubMed
-
- Kerr TP, Sewry CA, Robb SA, Roberts RG. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet. 2001;109:402–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

