Tissue enrichment analysis for C. elegans genomics
- PMID: 27618863
- PMCID: PMC5020436
- DOI: 10.1186/s12859-016-1229-9
Tissue enrichment analysis for C. elegans genomics
Abstract
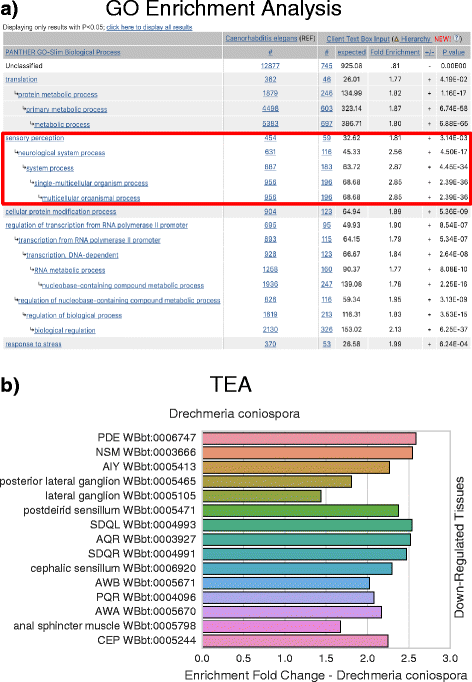
Background: Over the last ten years, there has been explosive development in methods for measuring gene expression. These methods can identify thousands of genes altered between conditions, but understanding these datasets and forming hypotheses based on them remains challenging. One way to analyze these datasets is to associate ontologies (hierarchical, descriptive vocabularies with controlled relations between terms) with genes and to look for enrichment of specific terms. Although Gene Ontology (GO) is available for Caenorhabditis elegans, it does not include anatomical information.
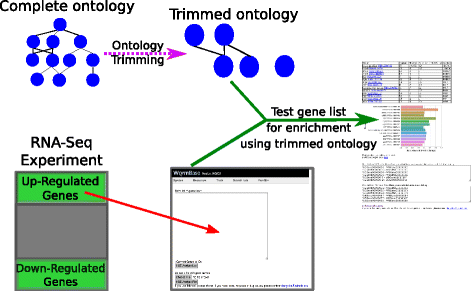
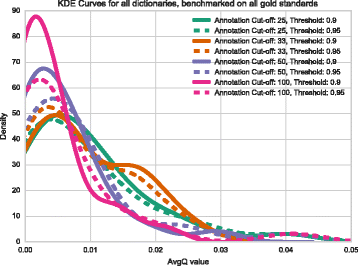
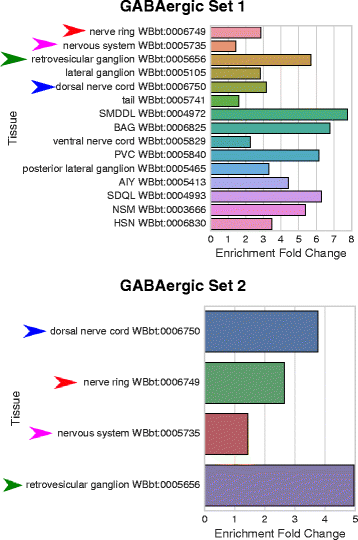
Results: We have developed a tool for identifying enrichment of C. elegans tissues among gene sets and generated a website GUI where users can access this tool. Since a common drawback to ontology enrichment analyses is its verbosity, we developed a very simple filtering algorithm to reduce the ontology size by an order of magnitude. We adjusted these filters and validated our tool using a set of 30 gold standards from Expression Cluster data in WormBase. We show our tool can even discriminate between embryonic and larval tissues and can even identify tissues down to the single-cell level. We used our tool to identify multiple neuronal tissues that are down-regulated due to pathogen infection in C. elegans.
Conclusions: Our Tissue Enrichment Analysis (TEA) can be found within WormBase, and can be downloaded using Python's standard pip installer. It tests a slimmed-down C. elegans tissue ontology for enrichment of specific terms and provides users with a text and graphic representation of the results.
Keywords: Anatomy ontology; Gene ontology; High-throughput biology; RNA-seq; WormBase.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

