A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve
- PMID: 27619409
- PMCID: PMC5027281
- DOI: 10.1038/ncomms12780
A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve
Abstract
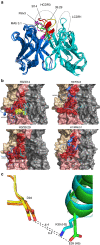
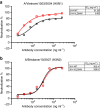
Understanding the natural evolution and structural changes involved in broadly neutralizing antibody (bnAb) development holds great promise for improving the design of prophylactic influenza vaccines. Here we report an haemagglutinin (HA) stem-directed bnAb, 3I14, isolated from human memory B cells, that utilizes a heavy chain encoded by the IGHV3-30 germline gene. MAb 3I14 binds and neutralizes groups 1 and 2 influenza A viruses and protects mice from lethal challenge. Analysis of VH and VL germline back-mutants reveals binding to H3 and H1 but not H5, which supports the critical role of somatic hypermutation in broadening the bnAb response. Moreover, a single VLD94N mutation improves the affinity of 3I14 to H5 by nearly 10-fold. These data provide evidence that memory B cell evolution can expand the HA subtype specificity. Our results further suggest that establishing an optimized memory B cell pool should be an aim of 'universal' influenza vaccine strategies.
Figures
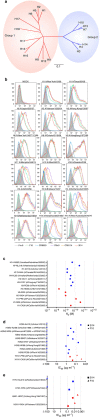
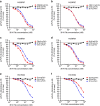
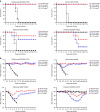

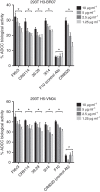
References
-
- WHO. WHO Influenza Fact Sheet No. 211, revised March 2003 (updated 19 October 2011). Available at http://www.who.int/mediacentre/factsheets/2003/fs211/en/ (2011).
-
- Nelson M. I. & Holmes E. C. The evolution of epidemic influenza. Nat. Rev. Genet. 8, 196–205 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

