Evolutionary mysteries in meiosis
- PMID: 27619705
- PMCID: PMC5031626
- DOI: 10.1098/rstb.2016.0001
Evolutionary mysteries in meiosis
Abstract
Meiosis is a key event of sexual life cycles in eukaryotes. Its mechanistic details have been uncovered in several model organisms, and most of its essential features have received various and often contradictory evolutionary interpretations. In this perspective, we present an overview of these often 'weird' features. We discuss the origin of meiosis (origin of ploidy reduction and recombination, two-step meiosis), its secondary modifications (in polyploids or asexuals, inverted meiosis), its importance in punctuating life cycles (meiotic arrests, epigenetic resetting, meiotic asymmetry, meiotic fairness) and features associated with recombination (disjunction constraints, heterochiasmy, crossover interference and hotspots). We present the various evolutionary scenarios and selective pressures that have been proposed to account for these features, and we highlight that their evolutionary significance often remains largely mysterious. Resolving these mysteries will likely provide decisive steps towards understanding why sex and recombination are found in the majority of eukaryotes.This article is part of the themed issue 'Weird sex: the underappreciated diversity of sexual reproduction'.
Keywords: automixis; epigenetics; genetic conflict; modified meiosis; origin of meiosis; recombination hotspots.
© 2016 The Author(s).
Figures

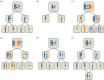
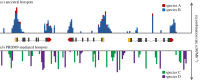
References
-
- Maynard Smith J. 1978. The evolution of sex. Cambridge, UK: Cambridge University Press.
-
- Hamilton WD. 2001. Narrow roads of gene land: vol. 2: evolution of sex. Oxford, UK: Oxford University Press.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

