A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1
- PMID: 27620479
- PMCID: PMC5075121
- DOI: 10.1128/AAC.01457-16
A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1
Abstract
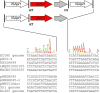
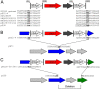
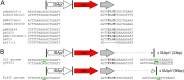
Analysis of mcr-1-containing sequences identified a common ∼2,607-bp DNA segment that in many cases is flanked on one or both ends by ISApl1 We present evidence that mcr-1 is mobilized by an ISApl1 composite transposon which has, in some cases, subsequently lost one or both copies of ISApl1 We also show that mcr-1 can be mobilized in some circumstances by a single upstream copy of ISApl1 in conjunction with the remnants of a downstream ISApl1.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161-168. doi:10.1016/S1473-3099(15)00424-7. - DOI - PubMed
-
- Zurfluh K, Klumpp J, Nuesch-Inderbinen M, Stephan R. 2016. Full-length nucleotide sequences of mcr-1 harboring plasmids isolated from extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli of different origins. Antimicrob Agents Chemother 60:5589–5591. doi:10.1128/AAC.00935-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

