Identification and Characterization of MCM3 as a Kelch-like ECH-associated Protein 1 (KEAP1) Substrate
- PMID: 27621311
- PMCID: PMC5095425
- DOI: 10.1074/jbc.M116.729418
Identification and Characterization of MCM3 as a Kelch-like ECH-associated Protein 1 (KEAP1) Substrate
Abstract
KEAP1 is a substrate adaptor protein for a CUL3-based E3 ubiquitin ligase. Ubiquitylation and degradation of the antioxidant transcription factor NRF2 is considered the primary function of KEAP1; however, few other KEAP1 substrates have been identified. Because KEAP1 is altered in a number of human pathologies and has been proposed as a potential therapeutic target therein, we sought to better understand KEAP1 through systematic identification of its substrates. Toward this goal, we combined parallel affinity capture proteomics and candidate-based approaches. Substrate-trapping proteomics yielded NRF2 and the related transcription factor NRF1 as KEAP1 substrates. Our targeted investigation of KEAP1-interacting proteins revealed MCM3, an essential subunit of the replicative DNA helicase, as a new substrate. We show that MCM3 is ubiquitylated by the KEAP1-CUL3-RBX1 complex in cells and in vitro Using ubiquitin remnant profiling, we identify the sites of KEAP1-dependent ubiquitylation in MCM3, and these sites are on predicted exposed surfaces of the MCM2-7 complex. Unexpectedly, we determined that KEAP1 does not regulate total MCM3 protein stability or subcellular localization. Our analysis of a KEAP1 targeting motif in MCM3 suggests that MCM3 is a point of direct contact between KEAP1 and the MCM hexamer. Moreover, KEAP1 associates with chromatin in a cell cycle-dependent fashion with kinetics similar to the MCM2-7 complex. KEAP1 is thus poised to affect MCM2-7 dynamics or function rather than MCM3 abundance. Together, these data establish new functions for KEAP1 within the nucleus and identify MCM3 as a novel substrate of the KEAP1-CUL3-RBX1 E3 ligase.
Keywords: DNA replication; E3 ubiquitin ligase; KEAP1; MCM3; NFE2L2; Nrf2; erythroid-derived 2-like factor; nuclear factor 2; oxidative stress; proteomics.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

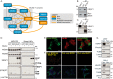

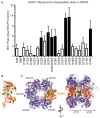


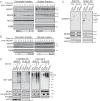
References
-
- Kobayashi A., Ohta T., and Yamamoto M. (2004) Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol. 378, 273–286 - PubMed
-
- Baird L., and Dinkova-Kostova A. T. (2013) Diffusion dynamics of the Keap1-Cullin3 interaction in single live cells. Biochem. Biophys. Res. Commun. 433, 58–65 - PubMed
-
- Jiang Z. Y., Chu H. X., Xi M. Y., Yang T. T., Jia J. M., Huang J. J., Guo X. K., Zhang X. J., You Q. D., and Sun H. P. (2013) Insight into the intermolecular recognition mechanism between Keap1 and IKKβ combining homology modelling, protein-protein docking, molecular dynamics simulations and virtual alanine mutation. PLoS ONE 8, e75076. - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

