Extracellular HSP110 skews macrophage polarization in colorectal cancer
- PMID: 27622020
- PMCID: PMC5006898
- DOI: 10.1080/2162402X.2016.1170264
Extracellular HSP110 skews macrophage polarization in colorectal cancer
Abstract
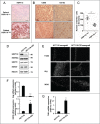
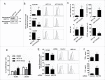
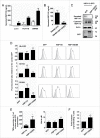
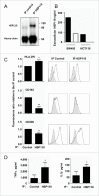
HSP110 is induced by different stresses and, through its anti-apoptotic and chaperoning properties, helps the cells to survive these adverse situations. In colon cancers, HSP110 is abnormally abundant. We have recently showed that colorectal cancer (CRC) patients with microsatellite instability (MSI) had an improved response to chemotherapy because they harbor an HSP110 inactivating mutation (HSP110DE9). In this work, we have used patients' biopsies and human CRC cells grown in vitro and in vivo (xenografts) to demonstrate that (1) HSP110 is secreted by CRC cells and that the amount of this extracellular HSP110 is strongly decreased by the expression of the mutant HSP110DE9, (2) Supernatants from CRC cells overexpressing HSP110 or purified recombinant human HSP110 (LPS-free) affect macrophage differentiation/polarization by favoring a pro-tumor, anti-inflammatory profile, (3) Conversely, inhibition of HSP110 (expression of siRNA, HSP110DE9 or immunodepletion) induced the formation of macrophages with a cytotoxic, pro-inflammatory profile. (4) Finally, this effect of extracellular HSP110 on macrophages seems to implicate TLR4. These results together with the fact that colorectal tumor biopsies with HSP110 high were infiltrated with macrophages with a pro-tumoral profile while those with HSP110 low were infiltrated with macrophages with a cytotoxic profile, suggest that the effect of extracellular HSP110 function on macrophages may also contribute to the poor outcomes associated with HSP110 expression.
Keywords: Cancer; colorectal; heat-shock protein; macrophage; polarization.
Figures
Similar articles
-
HSP110 promotes colorectal cancer growth through STAT3 activation.Oncogene. 2017 Apr 20;36(16):2328-2336. doi: 10.1038/onc.2016.403. Epub 2016 Nov 7. Oncogene. 2017. PMID: 27819670
-
Consequences of the Hsp110DE9 mutation in tumorigenesis and the 5-fluorouracil-based chemotherapy response in Msh2-deficient mice.Cell Mol Life Sci. 2022 Jun 1;79(6):332. doi: 10.1007/s00018-022-04293-3. Cell Mol Life Sci. 2022. PMID: 35648235 Free PMC article.
-
Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil–based chemotherapy.Gastroenterology. 2014 Feb;146(2):401-11.e1. doi: 10.1053/j.gastro.2013.10.054. Gastroenterology. 2014. PMID: 24512910
-
[Microsatellite instability and cancer: from genomic instability to personalized medicine].Med Sci (Paris). 2019 Jun-Jul;35(6-7):535-543. doi: 10.1051/medsci/2019093. Epub 2019 Jul 5. Med Sci (Paris). 2019. PMID: 31274083 Review. French.
-
Microsatellite instability: an update.Arch Toxicol. 2015 Jun;89(6):899-921. doi: 10.1007/s00204-015-1474-0. Epub 2015 Feb 22. Arch Toxicol. 2015. PMID: 25701956 Review.
Cited by
-
Heat Shock Proteins 27, 70, and 110: Expression and Prognostic Significance in Colorectal Cancer.Cancers (Basel). 2021 Aug 31;13(17):4407. doi: 10.3390/cancers13174407. Cancers (Basel). 2021. PMID: 34503216 Free PMC article.
-
The dark-side of the outside: how extracellular heat shock proteins promote cancer.Cell Mol Life Sci. 2021 May;78(9):4069-4083. doi: 10.1007/s00018-021-03764-3. Epub 2021 Feb 5. Cell Mol Life Sci. 2021. PMID: 33544155 Free PMC article. Review.
-
Chaperoning STAT3/5 by Heat Shock Proteins: Interest of Their Targeting in Cancer Therapy.Cancers (Basel). 2019 Dec 19;12(1):21. doi: 10.3390/cancers12010021. Cancers (Basel). 2019. PMID: 31861612 Free PMC article. Review.
-
Comprehensive analysis of heat shock proteins in glioma revealed the association with glioma-associated myeloid cells.Genes Immun. 2025 Jun;26(3):200-212. doi: 10.1038/s41435-025-00327-5. Epub 2025 Apr 15. Genes Immun. 2025. PMID: 40234584 Free PMC article.
-
Heat Shock Proteins in Lymphoma Immunotherapy.Front Immunol. 2021 Mar 18;12:660085. doi: 10.3389/fimmu.2021.660085. eCollection 2021. Front Immunol. 2021. PMID: 33815422 Free PMC article. Review.
References
-
- Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science 1993; 260:812-6; PMID:8484121; http://dx.doi.org/10.1126/science.8484121 - DOI - PubMed
-
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993; 363:558-61; PMID:8505985; http://dx.doi.org/10.1038/363558a0 - DOI - PubMed
-
- Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993; 260:816-9; PMID:8484122; http://dx.doi.org/10.1126/science.8484122 - DOI - PubMed
-
- Collura A, Lagrange A, Svrcek M, Marisa L, Buhard O, Guilloux A, Wanherdrick K, Dorard C, Taieb A, Saget A et al.. Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil-based chemotherapy. Gastroenterology 2014; 146:401-11 e1; PMID:24512910; http://dx.doi.org/10.1053/j.gastro.2013.10.054 - DOI - PubMed
-
- Duval A, Collura A, Berthenet K, Lagrange A, Garrido C. Microsatellite instability in colorectal cancer: time to stop hiding! Oncotarget 2011; 2:826-7; PMID:22084152; http://dx.doi.org/10.18632/oncotarget.353 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
