The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKε Kinase-Mediated Type-I IFN Antiviral Response
- PMID: 27622505
- PMCID: PMC5021333
- DOI: 10.1371/journal.ppat.1005880
The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKε Kinase-Mediated Type-I IFN Antiviral Response
Abstract
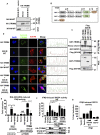
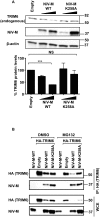
For efficient replication, viruses have developed mechanisms to evade innate immune responses, including the antiviral type-I interferon (IFN-I) system. Nipah virus (NiV), a highly pathogenic member of the Paramyxoviridae family (genus Henipavirus), is known to encode for four P gene-derived viral proteins (P/C/W/V) with IFN-I antagonist functions. Here we report that NiV matrix protein (NiV-M), which is important for virus assembly and budding, can also inhibit IFN-I responses. IFN-I production requires activation of multiple signaling components including the IκB kinase epsilon (IKKε). We previously showed that the E3-ubiquitin ligase TRIM6 catalyzes the synthesis of unanchored K48-linked polyubiquitin chains, which are not covalently attached to any protein, and activate IKKε for induction of IFN-I mediated antiviral responses. Using co-immunoprecipitation assays and confocal microscopy we show here that the NiV-M protein interacts with TRIM6 and promotes TRIM6 degradation. Consequently, NiV-M expression results in reduced levels of unanchored K48-linked polyubiquitin chains associated with IKKε leading to impaired IKKε oligomerization, IKKε autophosphorylation and reduced IFN-mediated responses. This IFN antagonist function of NiV-M requires a conserved lysine residue (K258) in the bipartite nuclear localization signal that is found in divergent henipaviruses. Consistent with this, the matrix proteins of Ghana, Hendra and Cedar viruses were also able to inhibit IFNβ induction. Live NiV infection, but not a recombinant NiV lacking the M protein, reduced the levels of endogenous TRIM6 protein expression. To our knowledge, matrix proteins of paramyxoviruses have never been reported to be involved in innate immune antagonism. We report here a novel mechanism of viral innate immune evasion by targeting TRIM6, IKKε and unanchored polyubiquitin chains. These findings expand the universe of viral IFN antagonism strategies and provide a new potential target for development of therapeutic interventions against NiV infections.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Medzhitov R, Preston-Hurlburt P, Janeway CA Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388(6640):394–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

