Reprogramming the Dynamin 2 mRNA by Spliceosome-mediated RNA Trans-splicing
- PMID: 27623444
- PMCID: PMC5056991
- DOI: 10.1038/mtna.2016.67
Reprogramming the Dynamin 2 mRNA by Spliceosome-mediated RNA Trans-splicing
Abstract
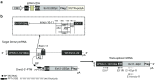
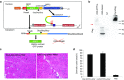
Dynamin 2 (DNM2) is a large GTPase, ubiquitously expressed, involved in membrane trafficking and regulation of actin and microtubule cytoskeletons. DNM2 mutations cause autosomal dominant centronuclear myopathy which is a rare congenital myopathy characterized by skeletal muscle weakness and histopathological features including nuclear centralization in absence of regeneration. No curative treatment is currently available for the DNM2-related autosomal dominant centronuclear myopathy. In order to develop therapeutic strategy, we evaluated here the potential of Spliceosome-Mediated RNA Trans-splicing technology to reprogram the Dnm2-mRNA in vitro and in vivo in mice. We show that classical 3'-trans-splicing strategy cannot be considered as accurate therapeutic strategy regarding toxicity of the pre-trans-splicing molecules leading to low rate of trans-splicing in vivo. Thus, we tested alternative strategies devoted to prevent this toxicity and enhance frequency of trans-splicing events. We succeeded to overcome the toxicity through a 5'-trans-splicing strategy which also allows detection of trans-splicing events at mRNA and protein levels in vitro and in vivo. These results suggest that the Spliceosome-Mediated RNA Trans-splicing strategy may be used to reprogram mutated Dnm2-mRNA but highlight the potential toxicity linked to the molecular tools which have to be carefully investigated during preclinical development.
Figures





References
-
- Durieux, AC, Prudhon, B, Guicheney, P and Bitoun, M (2010). Dynamin 2 and human diseases. J Mol Med (Berl) 88: 339–350. - PubMed
-
- Diatloff-Zito, C, Gordon, AJ, Duchaud, E and Merlin, G (1995). Isolation of an ubiquitously expressed cDNA encoding human dynamin II, a member of the large GTP-binding protein family. Gene 163: 301–306. - PubMed
-
- Jones, SM, Howell, KE, Henley, JR, Cao, H and McNiven, MA (1998). Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science 279: 573–577. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

