PARP10 deficiency manifests by severe developmental delay and DNA repair defect
- PMID: 27624574
- PMCID: PMC5096377
- DOI: 10.1007/s10048-016-0493-1
PARP10 deficiency manifests by severe developmental delay and DNA repair defect
Erratum in
-
Erratum to: PARP10 deficiency manifests by severe developmental delay and DNA repair defect.Neurogenetics. 2017 Apr;18(2):119. doi: 10.1007/s10048-017-0511-y. Neurogenetics. 2017. PMID: 28190220 No abstract available.
Abstract
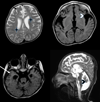
DNA repair mechanisms such as nucleotide excision repair (NER) and translesion synthesis (TLS) are dependent on proliferating cell nuclear antigen (PCNA), a DNA polymerase accessory protein. Recently, homozygosity for p.Ser228Ile mutation in the PCNA gene was reported in patients with neurodegeneration and impaired NER. Using exome sequencing, we identified a homozygous deleterious mutation, c.648delAG, in the PARP10 gene, in a patient suffering from severe developmental delay. In agreement, PARP10 protein was absent from the patient cells. We have previously shown that PARP10 is recruited by PCNA to DNA damage sites and is required for DNA damage resistance. The patient cells were significantly more sensitive to hydroxyurea and UV-induced DNA damage than control cells, resulting in increased apoptosis, indicating DNA repair impairment in the patient cells. PARP10 deficiency joins the long list of DNA repair defects associated with neurodegenerative disorders, including ataxia telangiectasia, xeroderma pigmentosum, Cockayne syndrome, and the recently reported PCNA mutation.
Keywords: DNA repair; Neurodegeneration.
Figures


References
-
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

