The PCP pathway regulates Baz planar distribution in epithelial cells
- PMID: 27624969
- PMCID: PMC5022056
- DOI: 10.1038/srep33420
The PCP pathway regulates Baz planar distribution in epithelial cells
Abstract
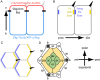
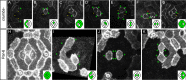
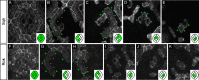
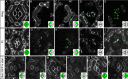
The localisation of apico-basal polarity proteins along the Z-axis of epithelial cells is well understood while their distribution in the plane of the epithelium is poorly characterised. Here we provide a systematic description of the planar localisation of apico-basal polarity proteins in the Drosophila ommatidial epithelium. We show that the adherens junction proteins Shotgun and Armadillo, as well as the baso-lateral complexes, are bilateral, i.e. present on both sides of cell interfaces. In contrast, we report that other key adherens junction proteins, Bazooka and the myosin regulatory light chain (Spaghetti squash) are unilateral, i.e. present on one side of cell interfaces. Furthermore, we demonstrate that planar cell polarity (PCP) and not the apical determinants Crumbs and Par-6 control Bazooka unilaterality in cone cells. Altogether, our work unravels an unexpected organisation and combination of apico-basal, cytoskeletal and planar polarity proteins that is different on either side of cell-cell interfaces and unique for the different contacts of the same cell.
Figures

References
-
- Tepass U., Theres C. & Knust E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 61, 787–799 (1990). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

