Combination of Whole Genome Sequencing, Linkage, and Functional Studies Implicates a Missense Mutation in Titin as a Cause of Autosomal Dominant Cardiomyopathy With Features of Left Ventricular Noncompaction
- PMID: 27625337
- PMCID: PMC5068189
- DOI: 10.1161/CIRCGENETICS.116.001431
Combination of Whole Genome Sequencing, Linkage, and Functional Studies Implicates a Missense Mutation in Titin as a Cause of Autosomal Dominant Cardiomyopathy With Features of Left Ventricular Noncompaction
Abstract
Background: High throughput next-generation sequencing techniques have made whole genome sequencing accessible in clinical practice; however, the abundance of variation in the human genomes makes the identification of a disease-causing mutation on a background of benign rare variants challenging.
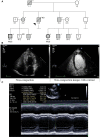
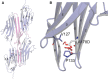
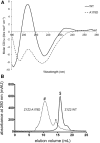
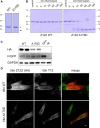
Methods and results: Here we combine whole genome sequencing with linkage analysis in a 3-generation family affected by cardiomyopathy with features of autosomal dominant left ventricular noncompaction cardiomyopathy. A missense mutation in the giant protein titin is the only plausible disease-causing variant that segregates with disease among the 7 surviving affected individuals, with interrogation of the entire genome excluding other potential causes. This A178D missense mutation, affecting a conserved residue in the second immunoglobulin-like domain of titin, was introduced in a bacterially expressed recombinant protein fragment and biophysically characterized in comparison to its wild-type counterpart. Multiple experiments, including size exclusion chromatography, small-angle x ray scattering, and circular dichroism spectroscopy suggest partial unfolding and domain destabilization in the presence of the mutation. Moreover, binding experiments in mammalian cells show that the mutation markedly impairs binding to the titin ligand telethonin.
Conclusions: Here we present genetic and functional evidence implicating the novel A178D missense mutation in titin as the cause of a highly penetrant familial cardiomyopathy with features of left ventricular noncompaction. This expands the spectrum of titin's roles in cardiomyopathies. It furthermore highlights that rare titin missense variants, currently often ignored or left uninterpreted, should be considered to be relevant for cardiomyopathies and can be identified by the approach presented here.
Keywords: cardiomyopathy; left ventricular noncompaction; missense mutation; telethonin; titin; whole genome sequencing.
© 2016 The Authors.
Figures
Comment in
-
Wrestling the Giant: New Approaches for Assessing Titin Variant Pathogenicity.Circ Cardiovasc Genet. 2016 Oct;9(5):392-394. doi: 10.1161/CIRCGENETICS.116.001594. Circ Cardiovasc Genet. 2016. PMID: 27756780 No abstract available.
-
Letter by Finsterer and Zarrouk-Mahjoub Regarding Article, "Combination of Whole Genome Sequencing, Linkage, and Functional Studies Implicates a Missense Mutation in Titin as a Cause of Autosomal Dominant Cardiomyopathy With Features of Left Ventricular Noncompaction".Circ Cardiovasc Genet. 2016 Dec;9(6):579. doi: 10.1161/CIRCGENETICS.116.001630. Circ Cardiovasc Genet. 2016. PMID: 27998947 No abstract available.
References
-
- Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N Engl J Med. 2011;364:1643–1656. doi: 10.1056/NEJMra0902923. - PubMed
-
- Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. - PubMed
-
- Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, et al. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123–1135a. doi: 10.1093/eurheartj/ehu301. - PubMed
-
- Watkins H. Assigning a causal role to genetic variants in hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2013;6:2–4. doi: 10.1161/CIRCGENETICS.111.000032. - PubMed
MeSH terms
Substances
Grants and funding
- MR/K015664/1/MRC_/Medical Research Council/United Kingdom
- RG/15/8/31480/BHF_/British Heart Foundation/United Kingdom
- MR/J010456/1/MRC_/Medical Research Council/United Kingdom
- CH/08/001/25300/BHF_/British Heart Foundation/United Kingdom
- PG/15/113/31944/BHF_/British Heart Foundation/United Kingdom
- RE/13/1/30181/BHF_/British Heart Foundation/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- RG/12/16/29939/BHF_/British Heart Foundation/United Kingdom
- 201543/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- FS/12/40/29712/BHF_/British Heart Foundation/United Kingdom
- NIHR-HCS-D13-04-006/DH_/Department of Health/United Kingdom

