Alternative Splicing, Internal Promoter, Nonsense-Mediated Decay, or All Three: Explaining the Distribution of Truncation Variants in Titin
- PMID: 27625338
- PMCID: PMC5068190
- DOI: 10.1161/CIRCGENETICS.116.001513
Alternative Splicing, Internal Promoter, Nonsense-Mediated Decay, or All Three: Explaining the Distribution of Truncation Variants in Titin
Abstract
Background: Truncating mutations in the giant sarcomeric gene Titin are the most common type of genetic alteration in dilated cardiomyopathy. Detailed studies have amassed a wealth of information about truncating variant position in cases and controls. Nonetheless, considerable confusion exists as to how to interpret the pathogenicity of these variants, hindering our ability to make useful recommendations to patients.
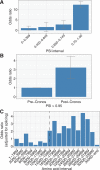
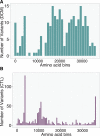
Methods and results: Building on our recent discovery of a conserved internal promoter within the Titin gene, we sought to develop an integrative statistical model to explain the observed pattern of Titin truncation variants in patients with dilated cardiomyopathy and population controls. We amassed Titin truncation mutation information from 1714 human dilated cardiomyopathy cases and >69 000 controls and found 3 factors explaining the distribution of Titin mutations: (1) alternative splicing, (2) whether the internal promoter Cronos isoform was disrupted, and (3) whether the distal C terminus was targeted (in keeping with the observation that truncation variants in this region escape nonsense-mediated decay and continue to be incorporated in the sarcomere). A model using these 3 factors had strong predictive performance with an area under the receiver operating characteristic curve of 0.81. Accordingly, individuals with either the most severe form of dilated cardiomyopathy or whose mutations demonstrated clear family segregation experienced the highest risk profile across all 3 components.
Conclusions: We conclude that quantitative models derived from large-scale human genetic and phenotypic data can be applied to help overcome the ever-growing challenges of genetic data interpretation. Results of our approach can be found at http://cvri.ucsf.edu/~deo/TTNtruncationvariant.html.
Keywords: alternative splicing; confusion; dilated cardiomyopathy; human; mutation.
© 2016 The Authors.
Figures


Comment in
-
Wrestling the Giant: New Approaches for Assessing Titin Variant Pathogenicity.Circ Cardiovasc Genet. 2016 Oct;9(5):392-394. doi: 10.1161/CIRCGENETICS.116.001594. Circ Cardiovasc Genet. 2016. PMID: 27756780 No abstract available.
References
-
- Gerull B, Gramlich M, Atherton J, McNabb M, Trombitás K, Sasse-Klaassen S, et al. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201–204. doi: 10.1038/ng815. - PubMed
-
- Itoh-Satoh M, Hayashi T, Nishi H, Koga Y, Arimura T, Koyanagi T, et al. Titin mutations as the molecular basis for dilated cardiomyopathy. Biochem Biophys Res Commun. 2002;291:385–393. doi: 10.1006/bbrc.2002.6448. - PubMed
-
- Gerull B, Atherton J, Geupel A, Sasse-Klaassen S, Heuser A, Frenneaux M, et al. Identification of a novel frameshift mutation in the giant muscle filament titin in a large Australian family with dilated cardiomyopathy. J Mol Med (Berl) 2006;84:478–483. doi: 10.1007/s00109-006-0060-6. - PubMed
-
- Carmignac V, Salih MA, Quijano-Roy S, Marchand S, Al Rayess MM, Mukhtar MM, et al. C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann Neurol. 2007;61:340–351. doi: 10.1002/ana.21089. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

