A Natural Light/Dark Cycle Regulation of Carbon-Nitrogen Metabolism and Gene Expression in Rice Shoots
- PMID: 27625675
- PMCID: PMC5003941
- DOI: 10.3389/fpls.2016.01318
A Natural Light/Dark Cycle Regulation of Carbon-Nitrogen Metabolism and Gene Expression in Rice Shoots
Abstract
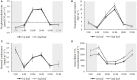
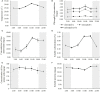
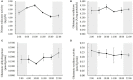
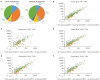
Light and temperature are two particularly important environmental cues for plant survival. Carbon and nitrogen are two essential macronutrients required for plant growth and development, and cellular carbon and nitrogen metabolism must be tightly coordinated. In order to understand how the natural light/dark cycle regulates carbon and nitrogen metabolism in rice plants, we analyzed the photosynthesis, key carbon-nitrogen metabolites, and enzyme activities, and differentially expressed genes and miRNAs involved in the carbon and nitrogen metabolic pathway in rice shoots at the following times: 2:00, 6:00, 10:00, 14:00, 18:00, and 22:00. Our results indicated that more CO2 was fixed into carbohydrates by a high net photosynthetic rate, respiratory rate, and stomatal conductance in the daytime. Although high levels of the nitrate reductase activity, free ammonium and carbohydrates were exhibited in the daytime, the protein synthesis was not significantly facilitated by the light and temperature. In mRNA sequencing, the carbon and nitrogen metabolism-related differentially expressed genes were obtained, which could be divided into eight groups: photosynthesis, TCA cycle, sugar transport, sugar metabolism, nitrogen transport, nitrogen reduction, amino acid metabolism, and nitrogen regulation. Additionally, a total of 78,306 alternative splicing events have been identified, which primarily belong to alternative 5' donor sites, alternative 3' acceptor sites, intron retention, and exon skipping. In sRNA sequencing, four carbon and nitrogen metabolism-related miRNAs (osa-miR1440b, osa-miR2876-5p, osa-miR1877 and osa-miR5799) were determined to be regulated by natural light/dark cycle. The expression level analysis showed that the four carbon and nitrogen metabolism-related miRNAs negatively regulated their target genes. These results may provide a good strategy to study how natural light/dark cycle regulates carbon and nitrogen metabolism to ensure plant growth and development.
Keywords: carbon; gene; metabolism; miRNA; nitrogen.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

