A highly invasive subpopulation of MDA-MB-231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo
- PMID: 27626697
- PMCID: PMC5356591
- DOI: 10.18632/oncotarget.11931
A highly invasive subpopulation of MDA-MB-231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo
Abstract
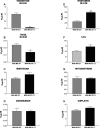
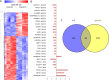
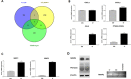
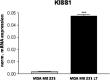
The acquisition of an invasive phenotype is a prerequisite for metastasization, yet it is not clear whether or to which extent the invasive phenotype is linked to other features characteristic of metastatic cells. We selected an invasive subpopulation from the triple negative breast cancer cell line MDA-MB-231, performing repeated cycles of preparative assays of invasion through Matrigel covered membranes. The invasive sub-population of MDA-MB-231 cells exhibits stronger migratory capacity as compared to parental cells confirming the highly invasive potential of the selected cell line. Prolonged cultivation of these cells did not abolish the invasive phenotype. ArrayCGH, DNA index quantification and karyotype analyses confirmed a common genetic origin of the parental and invasive subpopulations and revealed discrete structural differences of the invasive subpopulation including increased ploidy and the absence of a characteristic amplification of chromosome 5p14.1-15.33. Gene expression analyses showed a drastically altered expression profile including features of apocrine breast cancers and of invasion related matrix-metalloproteases and cytokines. The invasive cells showed accelerated proliferation, increased apoptosis, and an altered pattern of chemo-sensitivity with lower IC50 values for drugs affecting the mitotic apparatus. However, the invasive cell population is significantly less tumorigenic in orthotopic mouse xenografts suggesting that the acquisition of the invasive capacity and the achievement of metastatic growth potential are distinct events.
Keywords: aneuploidy; apocrine breast cancer; breast cancer; invasion; metastasis.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

References
-
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. - PubMed
-
- Allred DC, Mohsin SK, Fuqua SA. Histological and biological evolution of human premalignant breast disease. Endocr Relat Cancer. 2001;8:47–61. - PubMed
-
- Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van't Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65:9155–9158. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

