Characterization of a Wheat Breeders' Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum)
- PMID: 27627182
- PMCID: PMC5316916
- DOI: 10.1111/pbi.12635
Characterization of a Wheat Breeders' Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum)
Abstract
Targeted selection and inbreeding have resulted in a lack of genetic diversity in elite hexaploid bread wheat accessions. Reduced diversity can be a limiting factor in the breeding of high yielding varieties and crucially can mean reduced resilience in the face of changing climate and resource pressures. Recent technological advances have enabled the development of molecular markers for use in the assessment and utilization of genetic diversity in hexaploid wheat. Starting with a large collection of 819 571 previously characterized wheat markers, here we describe the identification of 35 143 single nucleotide polymorphism-based markers, which are highly suited to the genotyping of elite hexaploid wheat accessions. To assess their suitability, the markers have been validated using a commercial high-density Affymetrix Axiom® genotyping array (the Wheat Breeders' Array), in a high-throughput 384 microplate configuration, to characterize a diverse global collection of wheat accessions including landraces and elite lines derived from commercial breeding communities. We demonstrate that the Wheat Breeders' Array is also suitable for generating high-density genetic maps of previously uncharacterized populations and for characterizing novel genetic diversity produced by mutagenesis. To facilitate the use of the array by the wheat community, the markers, the associated sequence and the genotype information have been made available through the interactive web site 'CerealsDB'.
Keywords: genotyping array; single nucleotide polymorphism (SNP); wheat.
© 2016 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
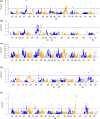
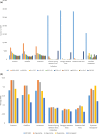
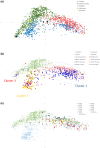

References
-
- Allen, A.M. , Barker, G.L. , Berry, S.T. , Coghill, J.A. , Gwilliam, R. , Kirby, S. , Robinson, P. , et al. (2011) Transcript‐specific, single‐nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 9, 1086–1099. - PubMed
-
- Allen, A.M. , Barker, G.L. , Wilkinson, P. , Burridge, A. , Winfield, M. , Coghill, J. , Uauy, C. , et al. (2013) Discovery and development of exome‐based, codominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol. J. 11, 279–295. - PubMed
-
- Brennan, J.P. and Quade, K.J. (2004) Analysis of the Impact of CIMMYT Research on the Australian Wheat Industry. Economic Research Report no. 25, NSW Department of Primary Industries, Wagga Wagga. Available from http://www.agric.nsw.gov.au/reader/10550.
-
- Burt, C. , Griffe, L.L. , Ridolfini, A.P. , Orford, S. , Griffiths, S. and Nicholson, P. (2014) Mining the Watkins collection of wheat landraces for novel sources of eyespot resistance. Plant. Pathol. 63, 1241–1250.
Publication types
MeSH terms
Grants and funding
- BB/L01386X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I002561/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/C/00005202/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/1002278/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

