Plasmablasts During Acute Dengue Infection Represent a Small Subset of a Broader Virus-specific Memory B Cell Pool
- PMID: 27628668
- PMCID: PMC5078588
- DOI: 10.1016/j.ebiom.2016.09.003
Plasmablasts During Acute Dengue Infection Represent a Small Subset of a Broader Virus-specific Memory B Cell Pool
Abstract
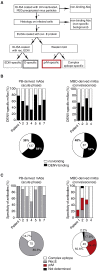
Dengue is endemic in tropical countries worldwide and the four dengue virus serotypes often co-circulate. Infection with one serotype results in high titers of cross-reactive antibodies produced by plasmablasts, protecting temporarily against all serotypes, but impairing protective immunity in subsequent infections. To understand the development of these plasmablasts, we analyzed virus-specific B cell properties in patients during acute disease and at convalescence. Plasmablasts were unrelated to classical memory cells expanding in the blood during early recovery. We propose that only a small subset of memory B cells is activated as plasmablasts during repeat infection and that plasmablast responses are not representative of the memory B cell repertoire after dengue infection.
Keywords: Antibodies; Dengue; Memory B cells; Plasmablasts.
Copyright © 2016 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- Halstead S.B., Rojanasuphot S., Sangkawibha N. Original antigenic sin in dengue. Am.J.Trop. Med. Hyg. 1983;32(1):154–156. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

