Generation of a Mutant Mucor hiemalis Endoglycosidase That Acts on Core-fucosylated N-Glycans
- PMID: 27629418
- PMCID: PMC5087746
- DOI: 10.1074/jbc.M116.737395
Generation of a Mutant Mucor hiemalis Endoglycosidase That Acts on Core-fucosylated N-Glycans
Abstract
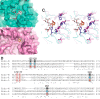
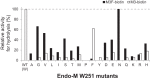
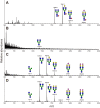
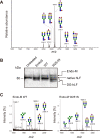
Endo-β-N-acetylglucosaminidase M (Endo-M), an endoglycosidase from the fungus Mucor hiemalis, is a useful tool for chemoenzymatic synthesis of glycoconjugates, including glycoprotein-based therapeutics having a precisely defined glycoform, by virtue of its transglycosylation activity. Although Endo-M has been known to act on various N-glycans, it does not act on core-fucosylated N-glycans, which exist widely in mammalian glycoproteins, thus limiting its application. Therefore, we performed site-directed mutagenesis on Endo-M to isolate mutant enzymes that are able to act on mammalian-type core-α1,6-fucosylated glycans. Among the Endo-M mutant enzymes generated, those in which the tryptophan at position 251 was substituted with alanine or asparagine showed altered substrate specificities. Such mutant enzymes exhibited increased hydrolysis of a synthetic α1,6-fucosylated trimannosyl core structure, whereas their activity on the afucosylated form decreased. In addition, among the Trp-251 mutants, the W251N mutant was most efficient in hydrolyzing the core-fucosylated substrate. W251N mutants could act on the immunoglobulin G-derived core-fucosylated glycopeptides and human lactoferrin glycoproteins. This mutant was also capable of transferring the sialyl glycan from an activated substrate intermediate (sialyl glyco-oxazoline) onto an α1,6-fucosyl-N-acetylglucosaminyl biotin. Furthermore, the W251N mutant gained a glycosynthase-like activity when a N175Q substitution was introduced and it caused accumulation of the transglycosylation products. These findings not only give insights into the substrate recognition mechanism of glycoside hydrolase family 85 enzymes but also widen their scope of application in preparing homogeneous glycoforms of core-fucosylated glycoproteins for the production of potent glycoprotein-based therapeutics.
Keywords: Endo-M; N-linked glycosylation; core fucose; endo-β-N-acetylglucosaminidase; glycoconjugate; glycoprotein; glycosidase; glycoside hydrolase; transglycosylation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Kato T., Kitamura K., Maeda M., Kimura Y., Katayama T., Ashida H., and Yamamoto K. (2007) Free oligosaccharides in the cytosol of Caenorhabditis elegans are generated through endoplasmic reticulum-Golgi trafficking. J. Biol. Chem. 282, 22080–22088 - PubMed
-
- Huang C., Harada Y., Hosomi A., Masahara-Negishi Y., Seino J., Fujihira H., Funakoshi Y., Suzuki T., Dohmae N., and Suzuki T. (2015) Endo-β-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proc. Natl. Acad. Sci. U.S.A. 112, 1398–1403 - PMC - PubMed
-
- Chantret I., and Moore S. E. H. (2008) Free oligosaccharide regulation during mammalian protein N-glycosylation. Glycobiology 18, 210–224 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

