An information model for computable cancer phenotypes
- PMID: 27629872
- PMCID: PMC5024416
- DOI: 10.1186/s12911-016-0358-4
An information model for computable cancer phenotypes
Abstract
Background: Standards, methods, and tools supporting the integration of clinical data and genomic information are an area of significant need and rapid growth in biomedical informatics. Integration of cancer clinical data and cancer genomic information poses unique challenges, because of the high volume and complexity of clinical data, as well as the heterogeneity and instability of cancer genome data when compared with germline data. Current information models of clinical and genomic data are not sufficiently expressive to represent individual observations and to aggregate those observations into longitudinal summaries over the course of cancer care. These models are acutely needed to support the development of systems and tools for generating the so called clinical "deep phenotype" of individual cancer patients, a process which remains almost entirely manual in cancer research and precision medicine.
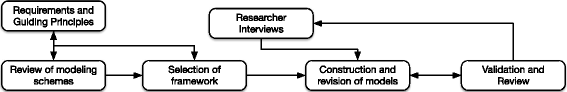
Methods: Reviews of existing ontologies and interviews with cancer researchers were used to inform iterative development of a cancer phenotype information model. We translated a subset of the Fast Healthcare Interoperability Resources (FHIR) models into the OWL 2 Description Logic (DL) representation, and added extensions as needed for modeling cancer phenotypes with terms derived from the NCI Thesaurus. Models were validated with domain experts and evaluated against competency questions.
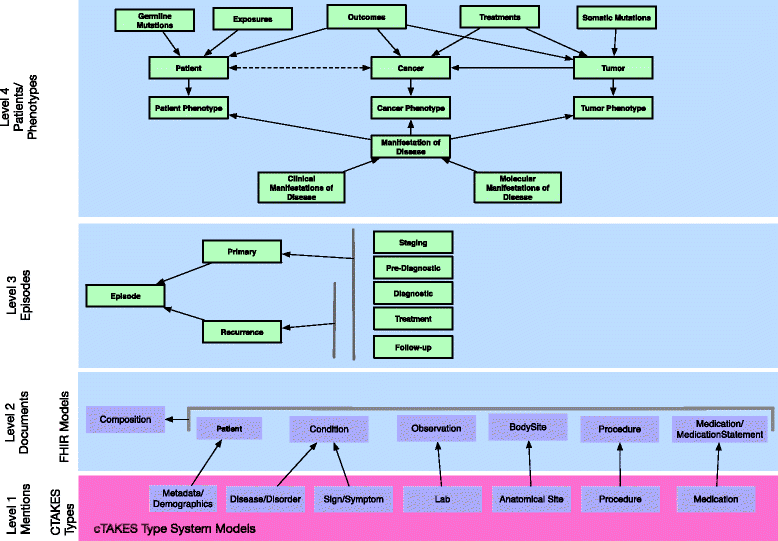
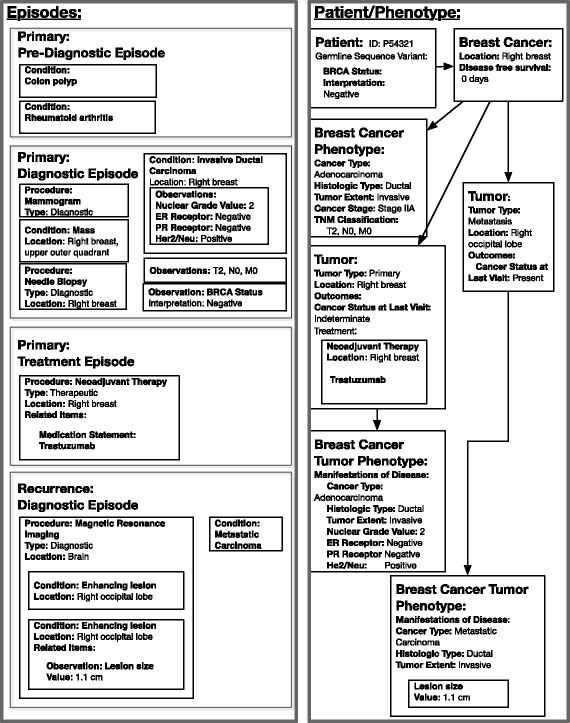
Results: The DeepPhe Information model represents cancer phenotype data at increasing levels of abstraction from mention level in clinical documents to summaries of key events and findings. We describe the model using breast cancer as an example, depicting methods to represent phenotypic features of cancers, tumors, treatment regimens, and specific biologic behaviors that span the entire course of a patient's disease.
Conclusions: We present a multi-scale information model for representing individual document mentions, document level classifications, episodes along a disease course, and phenotype summarization, linking individual observations to high-level summaries in support of subsequent integration and analysis.
Keywords: Cancer; Deep phenotyping; Information extraction; Information model.
Figures


References
-
- Index—FHIR v1.0.2 [http://hl7.org/fhir/]. Accessed 4 Sept 2016.
-
- Hiatt RA, Tai CG, Blayney DW, Deapen D, Hogarth M, Kizer KW, Lipscomb J, Malin J, Phillips SK, Santa J et al. Leveraging state cancer registries to measure and improve the quality of cancer care: a potential strategy for California and beyond. J Natl Cancer Inst 2015, 107 (5):djv047 - PubMed
-
- Helfand B, Roehl K, Cooper P, McGuire B, Fitzgerald L, Cancel-Tassin G, Cornu J-N, Bauer S, Van Blarigan E, Chen X et al. Associations of prostate cancer risk variants with disease aggressiveness: results of the NCI-SPORE Genetics Working Group analysis of 18,343 cases. Hum Genet. 2015;134(4):439–50. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

