Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology
- PMID: 27630250
- PMCID: PMC5116876
- DOI: 10.1128/MMBR.00003-16
Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology
Abstract
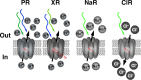
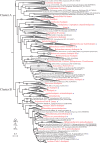
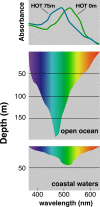
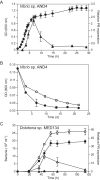
The recognition of a new family of rhodopsins in marine planktonic bacteria, proton-pumping proteorhodopsin, expanded the known phylogenetic range, environmental distribution, and sequence diversity of retinylidene photoproteins. At the time of this discovery, microbial ion-pumping rhodopsins were known solely in haloarchaea inhabiting extreme hypersaline environments. Shortly thereafter, proteorhodopsins and other light-activated energy-generating rhodopsins were recognized to be widespread among marine bacteria. The ubiquity of marine rhodopsin photosystems now challenges prior understanding of the nature and contributions of "heterotrophic" bacteria to biogeochemical carbon cycling and energy fluxes. Subsequent investigations have focused on the biophysics and biochemistry of these novel microbial rhodopsins, their distribution across the tree of life, evolutionary trajectories, and functional expression in nature. Later discoveries included the identification of proteorhodopsin genes in all three domains of life, the spectral tuning of rhodopsin variants to wavelengths prevailing in the sea, variable light-activated ion-pumping specificities among bacterial rhodopsin variants, and the widespread lateral gene transfer of biosynthetic genes for bacterial rhodopsins and their associated photopigments. Heterologous expression experiments with marine rhodopsin genes (and associated retinal chromophore genes) provided early evidence that light energy harvested by rhodopsins could be harnessed to provide biochemical energy. Importantly, some studies with native marine bacteria show that rhodopsin-containing bacteria use light to enhance growth or promote survival during starvation. We infer from the distribution of rhodopsin genes in diverse genomic contexts that different marine bacteria probably use rhodopsins to support light-dependent fitness strategies somewhere between these two extremes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

