Transcriptional and Physiological Changes during Mycobacterium tuberculosis Reactivation from Non-replicating Persistence
- PMID: 27630619
- PMCID: PMC5005354
- DOI: 10.3389/fmicb.2016.01346
Transcriptional and Physiological Changes during Mycobacterium tuberculosis Reactivation from Non-replicating Persistence
Abstract
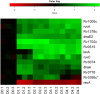
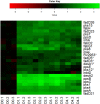
Mycobacterium tuberculosis can persist for years in the hostile environment of the host in a non-replicating or slowly replicating state. While active disease predominantly results from reactivation of a latent infection, the molecular mechanisms of M. tuberculosis reactivation are still poorly understood. We characterized the physiology and global transcriptomic profiles of M. tuberculosis during reactivation from hypoxia-induced non-replicating persistence. We found that M. tuberculosis reactivation upon reaeration was associated with a lag phase, in which the recovery of cellular physiological and metabolic functions preceded the resumption of cell replication. Enrichment analysis of the transcriptomic dynamics revealed changes to many metabolic pathways and transcription regulons/subnetworks that orchestrated the metabolic and physiological transformation in preparation for cell division. In particular, we found that M. tuberculosis reaeration lag phase is associated with down-regulation of persistence-associated regulons/subnetworks, including DosR, MprA, SigH, SigE, and ClgR, as well as metabolic pathways including those involved in the uptake of lipids and their catabolism. More importantly, we identified a number of up-regulated transcription regulons and metabolic pathways, including those involved in metal transport and remobilization, second messenger-mediated responses, DNA repair and recombination, and synthesis of major cell wall components. We also found that inactivation of the major alternative sigma factors SigE or SigH disrupted exit from persistence, underscoring the importance of the global transcriptional reprogramming during M. tuberculosis reactivation. Our observations suggest that M. tuberculosis lag phase is associated with a global gene expression reprogramming that defines the initiation of a reactivation process.
Keywords: RNA-Seq; gene expression profiling; lag phase; metabolism and physiology; reactivation; transcription regulon/subnetwork; tuberculosis.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

