Biochemical Characterization of Middle East Respiratory Syndrome Coronavirus Helicase
- PMID: 27631026
- PMCID: PMC5014916
- DOI: 10.1128/mSphere.00235-16
Biochemical Characterization of Middle East Respiratory Syndrome Coronavirus Helicase
Abstract
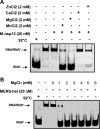
Middle East respiratory syndrome coronavirus (MERS-CoV) helicase is a superfamily 1 helicase containing seven conserved motifs. We have cloned, expressed, and purified a Strep-fused recombinant MERS-CoV nonstructural protein 13 (M-nsp13) helicase. Characterization of its biochemical properties showed that it unwound DNA and RNA similarly to severe acute respiratory syndrome CoV nsp13 (S-nsp13) helicase. We showed that M-nsp13 unwound in a 5'-to-3' direction and efficiently unwound the partially duplex RNA substrates with a long loading strand relative to those of the RNA substrates with a short or no loading strand. Moreover, the Km of ATP for M-nsp13 is inversely proportional to the length of the 5' loading strand of the partially duplex RNA substrates. Finally, we also showed that the rate of unwinding (ku) of M-nsp13 is directly proportional to the length of the 5' loading strand of the partially duplex RNA substrate. These results provide insights that enhance our understanding of the biochemical properties of M-nsp13. IMPORTANCE Coronaviruses are known to cause a wide range of diseases in humans and animals. Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel coronavirus discovered in 2012 and is responsible for acute respiratory syndrome in humans in the Middle East, Europe, North Africa, and the United States of America. Helicases are motor proteins that catalyze the processive separation of double-stranded nucleic acids into two single-stranded nucleic acids by utilizing the energy derived from ATP hydrolysis. MERS-CoV helicase is one of the most important viral replication enzymes of this coronavirus. Herein, we report the first bacterial expression, enzyme purification, and biochemical characterization of MERS-CoV helicase. The knowledge obtained from this study might be used to identify an inhibitor of MERS-CoV replication, and the helicase might be used as a therapeutic target.
Keywords: ATP hydrolysis; DNA; RNA; coronavirus; enzyme kinetics; helicase.
Figures








References
-
- Adedeji AO, Singh K, Kassim A, Coleman CM, Elliott R, Weiss SR, Frieman MB, Sarafianos SG. 2014. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother 58:4894–4898. doi:10.1128/AAC.02994-14. - DOI - PMC - PubMed
-
- Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi:10.1038/nature12005. - DOI - PMC - PubMed
-
- Hocke AC, Becher A, Knepper J, Peter A, Holland G, Tönnies M, Bauer TT, Schneider P, Neudecker J, Muth D, Wendtner CM, Rückert JC, Drosten C, Gruber AD, Laue M, Suttorp N, Hippenstiel S, Wolff T. 2013. Emerging human Middle East respiratory syndrome coronavirus causes widespread infection and alveolar damage in human lungs. Am J Respir Crit Care Med 188:882–886. doi:10.1164/rccm.201305-0954LE. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
