Exceptional Heterogeneity in Viral Evolutionary Dynamics Characterises Chronic Hepatitis C Virus Infection
- PMID: 27631086
- PMCID: PMC5025083
- DOI: 10.1371/journal.ppat.1005894
Exceptional Heterogeneity in Viral Evolutionary Dynamics Characterises Chronic Hepatitis C Virus Infection
Abstract
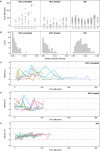
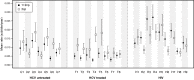
The treatment of HCV infection has seen significant progress, particularly since the approval of new direct-acting antiviral drugs. However these clinical achievements have been made despite an incomplete understanding of HCV replication and within-host evolution, especially compared with HIV-1. Here, we undertake a comprehensive analysis of HCV within-host evolution during chronic infection by investigating over 4000 viral sequences sampled longitudinally from 15 HCV-infected patients. We compare our HCV results to those from a well-studied HIV-1 cohort, revealing key differences in the evolutionary behaviour of these two chronic-infecting pathogens. Notably, we find an exceptional level of heterogeneity in the molecular evolution of HCV, both within and among infected individuals. Furthermore, these patterns are associated with the long-term maintenance of viral lineages within patients, which fluctuate in relative frequency in peripheral blood. Together, our findings demonstrate that HCV replication behavior is complex and likely comprises multiple viral subpopulations with distinct evolutionary dynamics. The presence of a structured viral population can explain apparent paradoxes in chronic HCV infection, such as rapid fluctuations in viral diversity and the reappearance of viral strains years after their initial detection.
Conflict of interest statement
RR is employed by BioInfExperts as bioinformatics consultant.
Figures


References
-
- Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, Yap PL, et al. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78 (Pt 2):321–8. - PubMed
-
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125(6):1808–17. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

