Identification of Interferon-Stimulated Genes with Antiretroviral Activity
- PMID: 27631702
- PMCID: PMC5026698
- DOI: 10.1016/j.chom.2016.08.005
Identification of Interferon-Stimulated Genes with Antiretroviral Activity
Abstract
Interferons (IFNs) exert their anti-viral effects by inducing the expression of hundreds of IFN-stimulated genes (ISGs). The activity of known ISGs is insufficient to account for the antiretroviral effects of IFN, suggesting that ISGs with antiretroviral activity are yet to be described. We constructed an arrayed library of ISGs from rhesus macaques and tested the ability of hundreds of individual macaque and human ISGs to inhibit early and late replication steps for 11 members of the retroviridae from various host species. These screens uncovered numerous ISGs with antiretroviral activity at both the early and late stages of virus replication. Detailed analyses of two antiretroviral ISGs indicate that indoleamine 2,3-dioxygenase 1 (IDO1) can inhibit retroviral replication by metabolite depletion while tripartite motif-56 (TRIM56) accentuates ISG induction by IFNα and inhibits the expression of late HIV-1 genes. Overall, these studies reveal numerous host proteins that mediate the antiretroviral activity of IFNs.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
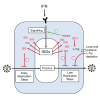




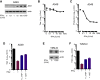


References
-
- Asmuth D.M., Murphy R.L., Rosenkranz S.L., Lertora J.J., Kottilil S., Cramer Y., Chan E.S., Schooley R.T., Rinaldo C.R., Thielman N., AIDS Clinical Trials Group A5192 Team Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa-2a in HIV-1-monoinfected participants: a phase II clinical trial. J. Infect. Dis. 2010;201:1686–1696. - PMC - PubMed
-
- Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S., Kazzaz Z., Bornstein E., Lambotte O., Altmann D. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006;12:1365–1371. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

