Characterization of the Specificity, Functionality, and Durability of Host T-Cell Responses Against the Full-Length Hepatitis E Virus
- PMID: 27631819
- PMCID: PMC5132006
- DOI: 10.1002/hep.28819
Characterization of the Specificity, Functionality, and Durability of Host T-Cell Responses Against the Full-Length Hepatitis E Virus
Abstract
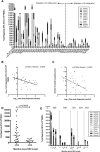
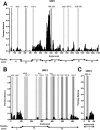
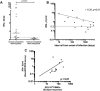
The interplay between host antiviral immunity and immunopathology during hepatitis E virus (HEV) infection determines important clinical outcomes. We characterized the specificity, functionality, and durability of host T-cell responses against the full-length HEV virus and assessed a novel "Quantiferon" assay for the rapid diagnosis of HEV infection. Eighty-nine volunteers were recruited from Oxford, Truro (UK), and Toulouse (France), including 44 immune-competent patients with acute HEV infection, 18 HEV-exposed immunosuppressed organ-transplant recipients (8 with chronic HEV), and 27 healthy volunteers. A genotype 3a peptide library (616 overlapping peptides spanning open reading frames [ORFs] 1-3) was used in interferon-gamma (IFN-γ) T-cell ELISpot assays. CD4+ /CD8+ T-cell subsets and polyfunctionality were defined using ICCS and SPICE analysis. Quantification of IFN-γ used whole-blood stimulation with recombinant HEV-capsid protein in the QuantiFERON kit. HEV-specific T-cell responses were detected in 41/44 immune-competent HEV exposed volunteers (median magnitude: 397 spot-forming units/106 peripheral blood mononuclear cells), most frequently targeting ORF2. High-magnitude, polyfunctional CD4 and CD8+ T cells were detected during acute disease and maintained to 12 years, but these declined over time, with CD8+ responses becoming more monofunctional. Low-level responses were detectable in immunosuppressed patients. Twenty-three novel HEV CD4+ and CD8+ T-cell targets were mapped predominantly to conserved genomic regions. QuantiFERON testing demonstrated an inverse correlation between IFN-γ production and the time from clinical presentation, providing 100% specificity, and 71% sensitivity (area under the receiver operator characteristic curve of 0.86) for HEV exposure at 0.3 IU/mL.
Conclusion: Robust HEV-specific T-cell responses generated during acute disease predominantly target ORF2, but decline in magnitude and polyfunctionality over time. Defining HEV T-cell targets will be important for the investigation of HEV-associated autoimmune disease. (Hepatology 2016;64:1934-1950).
© 2016 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of the American Association for the Study of Liver Diseases.
Figures



References
-
- Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 2012;55:988–997. - PubMed
-
- Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 2008;8:698–709. - PubMed
-
- Khuroo MS. Discovery of hepatitis E: the epidemic non‐A, non‐B hepatitis 30 years down the memory lane. Virus Res 2011;161:3–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

