3D Cell Culture in a Self-Assembled Nanofiber Environment
- PMID: 27632425
- PMCID: PMC5025053
- DOI: 10.1371/journal.pone.0162853
3D Cell Culture in a Self-Assembled Nanofiber Environment
Abstract
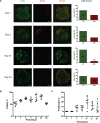
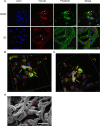
The development and utilization of three-dimensional cell culture platforms has been gaining more traction. Three-dimensional culture platforms are capable of mimicking in vivo microenvironments, which provide greater physiological relevance in comparison to conventional two-dimensional cultures. The majority of three-dimensional culture platforms are challenged by the lack of cell attachment, long polymerization times, and inclusion of undefined xenobiotics, and cytotoxic cross-linkers. In this study, we review the use of a highly defined material composed of naturally occurring compounds, hyaluronic acid and chitosan, known as Cell-Mate3DTM. Moreover, we provide an original measurement of Young's modulus using a uniaxial unconfined compression method to elucidate the difference in microenvironment rigidity for acellular and cellular conditions. When hydrated into a tissue-like hybrid hydrocolloid/hydrogel, Cell-Mate3DTM is a highly versatile three-dimensional culture platform that enables downstream applications such as flow cytometry, immunostaining, histological staining, and functional studies to be applied with relative ease.
Conflict of interest statement
JHB is the inventor of Cell—Mate3D and is a shareholder and the founder, Chairman of the Board of Directors and Chief Science Officer of Bioactive Regenerative Therapeutics, Inc. (BRTI) Life Sciences, and YC, EL and JDG are employees of BRTI Life Sciences. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures




References
-
- Nema R, Khare S. An animal cell culture: Advance technology for modern research. Advances in Bioscience and Biotechnology 2012;3:219–26. 10.4236/abb.2012.33030 - DOI
-
- Bissell MJ, Barcellos-Hoff MH. The influence of extracellular matrix on gene expression: is structure the message? Journal of Cell Science Supplement 1987;8:327–43. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

