Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos
- PMID: 27633012
- PMCID: PMC5066237
- DOI: 10.1101/gad.285767.116
Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos
Abstract
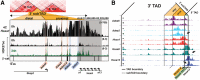
Sequential 3'-to-5' activation of the Hox gene clusters in early embryos is a most fascinating issue in developmental biology. Neither the trigger nor the regulatory elements involved in the transcriptional initiation of the 3'-most Hox genes have been unraveled in any organism. We demonstrate that a series of enhancers, some of which are Wnt-dependent, is located within a HoxA 3' subtopologically associated domain (subTAD). This subTAD forms the structural basis for multiple layers of 3'-polarized features, including DNA accessibility and enhancer activation. Deletion of the cassette of Wnt-dependent enhancers proves its crucial role in initial transcription of HoxA at the 3' side of the cluster.
Keywords: DNA accessibility; Hox regulation; chromatin conformation; developmental enhancers; regulatory landscapes.
© 2016 Neijts et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
