Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp
- PMID: 27633015
- PMCID: PMC5066242
- DOI: 10.1101/gad.287094.116
Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp
Abstract
In mammals, body temperature fluctuates diurnally around a mean value of 36°C-37°C. Despite the small differences between minimal and maximal values, body temperature rhythms can drive robust cycles in gene expression in cultured cells and, likely, animals. Here we studied the mechanisms responsible for the temperature-dependent expression of cold-inducible RNA-binding protein (CIRBP). In NIH3T3 fibroblasts exposed to simulated mouse body temperature cycles, Cirbp mRNA oscillates about threefold in abundance, as it does in mouse livers. This daily mRNA accumulation cycle is directly controlled by temperature oscillations and does not depend on the cells' circadian clocks. Here we show that the temperature-dependent accumulation of Cirbp mRNA is controlled primarily by the regulation of splicing efficiency, defined as the fraction of Cirbp pre-mRNA processed into mature mRNA. As revealed by genome-wide "approach to steady-state" kinetics, this post-transcriptional mechanism is widespread in the temperature-dependent control of gene expression.
Keywords: Cirbp; circadian rhythms; splicing efficiency; temperature.
© 2016 Gotic et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
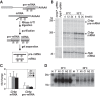

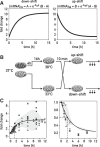

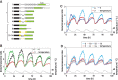
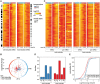
 and × symbols in downshift and * and × symbols in upshift experiments in Fig. 3C) are shown for each temperature shift. Gene structures are depicted below the tracks. (B,C) Quantification of Midn and Cirbp mRNA and pre-mRNA expression for the two biological replicate series/temperature shift. Black circles and green triangles represent the RNA read counts (expressed in RPKM [reads per kilobase per million mapped reads]) for mRNA and pre-mRNA, respectively. Data points indicated by the infinity symbol on the X-axis correspond to the expression at t0 from the opposite temperature shift and are thus assumed to reflect the opposite steady state. Solid black and green lines represent the model predictions for mRNA and pre-mRNA accumulation. The shaded areas indicate SDs (details are in the
and × symbols in downshift and * and × symbols in upshift experiments in Fig. 3C) are shown for each temperature shift. Gene structures are depicted below the tracks. (B,C) Quantification of Midn and Cirbp mRNA and pre-mRNA expression for the two biological replicate series/temperature shift. Black circles and green triangles represent the RNA read counts (expressed in RPKM [reads per kilobase per million mapped reads]) for mRNA and pre-mRNA, respectively. Data points indicated by the infinity symbol on the X-axis correspond to the expression at t0 from the opposite temperature shift and are thus assumed to reflect the opposite steady state. Solid black and green lines represent the model predictions for mRNA and pre-mRNA accumulation. The shaded areas indicate SDs (details are in the 
Comment in
-
'Cold cuts' added to the circadian smorgasbord of regulatory mechanisms.Genes Dev. 2016 Sep 1;30(17):1909-10. doi: 10.1101/gad.289587.116. Genes Dev. 2016. PMID: 27664233 Free PMC article.
References
-
- Adjibade P, Mazroui R. 2014. Control of mRNA turnover: implication of cytoplasmic RNA granules. Semin Cell Dev Biol 34: 15–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
