Integrative Genomic Analysis Identifies the Core Transcriptional Hallmarks of Human Hepatocellular Carcinoma
- PMID: 27634755
- PMCID: PMC5660733
- DOI: 10.1158/0008-5472.CAN-16-1559
Integrative Genomic Analysis Identifies the Core Transcriptional Hallmarks of Human Hepatocellular Carcinoma
Abstract
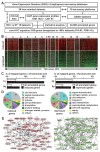
Integrative genomics helped characterize molecular heterogeneity in hepatocellular carcinoma (HCC), leading to targeted drug candidates for specific HCC subtypes. However, no consensus was achieved for genes and pathways commonly altered in HCC. Here, we performed a meta-analysis of 15 independent datasets (n = 784 human HCC) and identified a comprehensive signature consisting of 935 genes commonly deregulated in HCC as compared with the surrounding nontumor tissue. In the HCC signature, upregulated genes were linked to early genomic alterations in hepatocarcinogenesis, particularly gains of 1q and 8q. The HCC signature covered well-established cancer hallmarks, such as proliferation, metabolic reprogramming, and microenvironment remodeling, together with specific hallmarks associated with protein turnover and epigenetics. Subsequently, the HCC signature enabled us to assess the efficacy of signature-relevant drug candidates, including histone deacetylase inhibitors that specifically reduced the viability of six human HCC cell lines. Overall, this integrative genomics approach identified cancer hallmarks recurrently altered in human HCC that may be targeted by specific drugs. Combined therapies targeting common and subtype-specific cancer networks may represent a relevant therapeutic strategy in liver cancer. Cancer Res; 76(21); 6374-81. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures


References
-
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31(4):339–46. - PubMed
-
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Transancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46(12):1267–73. - PubMed
-
- Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15(11):653–67. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

