Cell Adhesion Molecule CD166 Drives Malignant Progression and Osteolytic Disease in Multiple Myeloma
- PMID: 27634757
- PMCID: PMC5135585
- DOI: 10.1158/0008-5472.CAN-16-0517
Cell Adhesion Molecule CD166 Drives Malignant Progression and Osteolytic Disease in Multiple Myeloma
Abstract
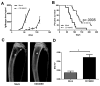
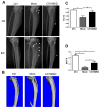
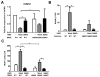
Multiple myeloma is incurable once osteolytic lesions have seeded at skeletal sites, but factors mediating this deadly pathogenic advance remain poorly understood. Here, we report evidence of a major role for the cell adhesion molecule CD166, which we discovered to be highly expressed in multiple myeloma cell lines and primary bone marrow cells from patients. CD166+ multiple myeloma cells homed more efficiently than CD166- cells to the bone marrow of engrafted immunodeficient NSG mice. CD166 silencing in multiple myeloma cells enabled longer survival, a smaller tumor burden, and less osteolytic lesions, as compared with mice bearing control cells. CD166 deficiency in multiple myeloma cell lines or CD138+ bone marrow cells from multiple myeloma patients compromised their ability to induce bone resorption in an ex vivo organ culture system. Furthermore, CD166 deficiency in multiple myeloma cells also reduced the formation of osteolytic disease in vivo after intratibial engraftment. Mechanistic investigation revealed that CD166 expression in multiple myeloma cells inhibited osteoblastogenesis of bone marrow-derived osteoblast progenitors by suppressing Runx2 gene expression. Conversely, CD166 expression in multiple myeloma cells promoted osteoclastogenesis by activating TRAF6-dependent signaling pathways in osteoclast progenitors. Overall, our results define CD166 as a pivotal director in multiple myeloma cell homing to the bone marrow and multiple myeloma progression, rationalizing its further study as a candidate therapeutic target for multiple myeloma treatment. Cancer Res; 76(23); 6901-10. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Slovak ML. Multiple myeloma: current perspectives. Clinics in laboratory medicine. 2011;31(4):699–724. x. - PubMed
-
- Mitsiades CS, McMillin DW, Klippel S, Hideshima T, Chauhan D, Richardson PG, et al. The role of the bone marrow microenvironment in the pathophysiology of myeloma and its significance in the development of more effective therapies. Hematology/oncology clinics of North America. 2007;21(6):1007–34. vii–viii. - PubMed
-
- Van Camp B, Van Riet I. Homing mechanisms in the biology of multiple myeloma. Verhandelingen - Koninklijke Academie voor Geneeskunde van Belgie. 1998;60(3):163–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

