Co-regulation of paralog genes in the three-dimensional chromatin architecture
- PMID: 27634932
- PMCID: PMC5224500
- DOI: 10.1093/nar/gkw813
Co-regulation of paralog genes in the three-dimensional chromatin architecture
Abstract
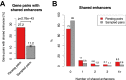
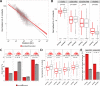
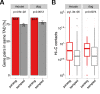
Paralog genes arise from gene duplication events during evolution, which often lead to similar proteins that cooperate in common pathways and in protein complexes. Consequently, paralogs show correlation in gene expression whereby the mechanisms of co-regulation remain unclear. In eukaryotes, genes are regulated in part by distal enhancer elements through looping interactions with gene promoters. These looping interactions can be measured by genome-wide chromatin conformation capture (Hi-C) experiments, which revealed self-interacting regions called topologically associating domains (TADs). We hypothesize that paralogs share common regulatory mechanisms to enable coordinated expression according to TADs. To test this hypothesis, we integrated paralogy annotations with human gene expression data in diverse tissues, genome-wide enhancer-promoter associations and Hi-C experiments in human, mouse and dog genomes. We show that paralog gene pairs are enriched for co-localization in the same TAD, share more often common enhancer elements than expected and have increased contact frequencies over large genomic distances. Combined, our results indicate that paralogs share common regulatory mechanisms and cluster not only in the linear genome but also in the three-dimensional chromatin architecture. This enables concerted expression of paralogs over diverse cell-types and indicate evolutionary constraints in functional genome organization.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Koonin E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. - PubMed
-
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. - PubMed
-
- Carter D., Chakalova L., Osborne C.S., Dai Y.F., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

