Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division
- PMID: 27635761
- PMCID: PMC5028122
- DOI: 10.1016/j.molcel.2016.08.013
Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division
Abstract
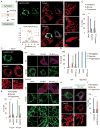
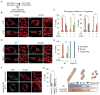
Mitochondria divide to control their size, distribution, turnover, and function. Dynamin-related protein 1 (Drp1) is a critical mechanochemical GTPase that drives constriction during mitochondrial division. It is generally believed that mitochondrial division is regulated during recruitment of Drp1 to mitochondria and its oligomerization into a division apparatus. Here, we report an unforeseen mechanism that regulates mitochondrial division by coincident interactions of Drp1 with the head group and acyl chains of phospholipids. Drp1 recognizes the head group of phosphatidic acid (PA) and two saturated acyl chains of another phospholipid by penetrating into the hydrophobic core of the membrane. The dual phospholipid interactions restrain Drp1 via inhibition of oligomerization-stimulated GTP hydrolysis that promotes membrane constriction. Moreover, a PA-producing phospholipase, MitoPLD, binds Drp1, creating a PA-rich microenvironment in the vicinity of a division apparatus. Thus, PA controls the activation of Drp1 after the formation of the division apparatus.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Restraining the Divider: A Drp1-Phospholipid Interaction Inhibits Drp1 Activity and Shifts the Balance from Mitochondrial Fission to Fusion.Mol Cell. 2016 Sep 15;63(6):913-5. doi: 10.1016/j.molcel.2016.08.033. Mol Cell. 2016. PMID: 27635756
References
-
- Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

