Solution structure of protein synthesis initiation factor 1 from Pseudomonas aeruginosa
- PMID: 27636899
- PMCID: PMC5119568
- DOI: 10.1002/pro.3042
Solution structure of protein synthesis initiation factor 1 from Pseudomonas aeruginosa
Abstract
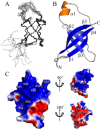
Pseudomonas aeruginosa is an opportunistic bacterial pathogen and a primary cause of nosocomial infection in humans. The rate of antibiotic resistance in P. aeruginosa is increasing worldwide leading to an unmet need for discovery of new chemical compounds distinctly different from present antimicrobials. Protein synthesis is an essential metabolic process and a validated target for the development of new antibiotics. Initiation factor 1 from P. aeruginosa (Pa-IF1) is the smallest of the three initiation factors that act to establish the 30S initiation complex during initiation of protein biosynthesis. Here we report the characterization and solution NMR structure of Pa-IF1. Pa-IF1 consists of a five-stranded β-sheet with an unusual extended β-strand at the C-terminus and one short α-helix arranged in the sequential order β1-β2-β3-α1-β4-β5. The structure adopts a typical β-barrel fold and contains an oligomer-binding motif. A cluster of basic residues (K39, R41, K42, K64, R66, R70, and R72) located on the surface of strands β4 and β5 near the short α-helix may compose the binding interface with the 30S subunit.
Keywords: NMR structure; Pseudomonas aeruginosa; translation initiation factor 1; β-barrel motif.
© 2016 The Protein Society.
Figures

References
-
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock‐Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. - PubMed
-
- McCoy LS, Xie Y, Tor Y (2011) Antibiotics that target protein synthesis. RNA 2:209–232. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

