A Variable Genetic Architecture of Melanic Evolution in Drosophila melanogaster
- PMID: 27638419
- PMCID: PMC5105859
- DOI: 10.1534/genetics.116.192492
A Variable Genetic Architecture of Melanic Evolution in Drosophila melanogaster
Abstract
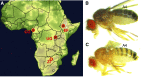
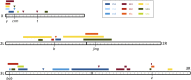
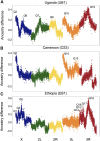
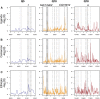
Unraveling the genetic architecture of adaptive phenotypic divergence is a fundamental quest in evolutionary biology. In Drosophila melanogaster, high-altitude melanism has evolved in separate mountain ranges in sub-Saharan Africa, potentially as an adaptation to UV intensity. We investigated the genetic basis of this melanism in three populations using a new bulk segregant analysis mapping method. We identified 19 distinct QTL regions from nine mapping crosses, with several QTL peaks overlapping between two or all populations, and yet different crosses involving the same melanic population commonly yielded distinct QTL. The strongest QTL often overlapped well-known pigmentation genes, but we typically did not find wide signals of genetic differentiation (FST) between lightly and darkly pigmented populations at these genes. Instead, we found small numbers of highly differentiated SNPs at the probable causative genes. A simulation analysis showed that these patterns of polymorphism were consistent with selection on standing genetic variation. Overall, our results suggest that, even for potentially simpler traits like pigmentation, the complexity of adaptive trait evolution poses important challenges for QTL mapping and population genetic analysis.
Keywords: Drosophila melanogaster; adaptation; bulk segregant analysis; parallel evolution; pigmentation; standing genetic variation.
Copyright © 2016 Bastide et al.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

