Alkylating Agent-Induced NRF2 Blocks Endoplasmic Reticulum Stress-Mediated Apoptosis via Control of Glutathione Pools and Protein Thiol Homeostasis
- PMID: 27638861
- PMCID: PMC5136348
- DOI: 10.1158/1535-7163.MCT-16-0271
Alkylating Agent-Induced NRF2 Blocks Endoplasmic Reticulum Stress-Mediated Apoptosis via Control of Glutathione Pools and Protein Thiol Homeostasis
Abstract
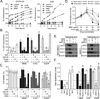
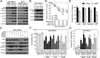
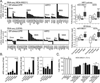
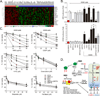
Alkylating agents are a commonly used cytotoxic class of anticancer drugs. Understanding the mechanisms whereby cells respond to these drugs is key to identify means to improve therapy while reducing toxicity. By integrating genome-wide gene expression profiling, protein analysis, and functional cell validation, we herein demonstrated a direct relationship between NRF2 and Endoplasmic Reticulum (ER) stress pathways in response to alkylating agents, which is coordinated by the availability of glutathione (GSH) pools. GSH is essential for both drug detoxification and protein thiol homeostasis within the ER, thus inhibiting ER stress induction and promoting survival, an effect independent of its antioxidant role. NRF2 accumulation induced by alkylating agents resulted in increased GSH synthesis via GCLC/GCLM enzyme, and interfering with this NRF2 response by either NRF2 knockdown or GCLC/GCLM inhibition with buthionine sulfoximine caused accumulation of damaged proteins within the ER, leading to PERK-dependent apoptosis. Conversely, upregulation of NRF2, through KEAP1 depletion or NRF2-myc overexpression, or increasing GSH levels with N-acetylcysteine or glutathione-ethyl-ester, decreased ER stress and abrogated alkylating agents-induced cell death. Based on these results, we identified a subset of lung and head-and-neck carcinomas with mutations in either KEAP1 or NRF2/NFE2L2 genes that correlate with NRF2 target overexpression and poor survival. In KEAP1-mutant cancer cells, NRF2 knockdown and GSH depletion increased cell sensitivity via ER stress induction in a mechanism specific to alkylating drugs. Overall, we show that the NRF2-GSH influence on ER homeostasis implicates defects in NRF2-GSH or ER stress machineries as affecting alkylating therapy toxicity. Mol Cancer Ther; 15(12); 3000-14. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors declare that no conflict of interest exists.
Figures



References
-
- Hernandez-Aya LF, Gonzalez-Angulo AM. Adjuvant systemic therapies in breast cancer. Surg Clin North Am. 2013;93:473–491. - PubMed
-
- Jones RB, Matthes S, Dufton C, Bearman SI, Stemmer SM, Meyers S, et al. Pharmacokinetic/pharmacodynamic interactions of intensive cyclophosphamide, cisplatin, and BCNU in patients with breast cancer. Breast Cancer Res Treat. 1993;26(Suppl):S11–S17. - PubMed
-
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. - PubMed
-
- Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6:2585–2597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

