Metabolomics enables precision medicine: "A White Paper, Community Perspective"
- PMID: 27642271
- PMCID: PMC5009152
- DOI: 10.1007/s11306-016-1094-6
Metabolomics enables precision medicine: "A White Paper, Community Perspective"
Abstract
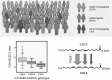
Introduction background to metabolomics: Metabolomics is the comprehensive study of the metabolome, the repertoire of biochemicals (or small molecules) present in cells, tissues, and body fluids. The study of metabolism at the global or "-omics" level is a rapidly growing field that has the potential to have a profound impact upon medical practice. At the center of metabolomics, is the concept that a person's metabolic state provides a close representation of that individual's overall health status. This metabolic state reflects what has been encoded by the genome, and modified by diet, environmental factors, and the gut microbiome. The metabolic profile provides a quantifiable readout of biochemical state from normal physiology to diverse pathophysiologies in a manner that is often not obvious from gene expression analyses. Today, clinicians capture only a very small part of the information contained in the metabolome, as they routinely measure only a narrow set of blood chemistry analytes to assess health and disease states. Examples include measuring glucose to monitor diabetes, measuring cholesterol and high density lipoprotein/low density lipoprotein ratio to assess cardiovascular health, BUN and creatinine for renal disorders, and measuring a panel of metabolites to diagnose potential inborn errors of metabolism in neonates.
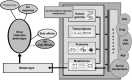
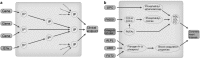
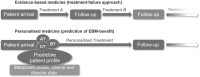
Objectives of white paper—expected treatment outcomes and metabolomics enabling tool for precision medicine: We anticipate that the narrow range of chemical analyses in current use by the medical community today will be replaced in the future by analyses that reveal a far more comprehensive metabolic signature. This signature is expected to describe global biochemical aberrations that reflect patterns of variance in states of wellness, more accurately describe specific diseases and their progression, and greatly aid in differential diagnosis. Such future metabolic signatures will: (1) provide predictive, prognostic, diagnostic, and surrogate markers of diverse disease states; (2) inform on underlying molecular mechanisms of diseases; (3) allow for sub-classification of diseases, and stratification of patients based on metabolic pathways impacted; (4) reveal biomarkers for drug response phenotypes, providing an effective means to predict variation in a subject's response to treatment (pharmacometabolomics); (5) define a metabotype for each specific genotype, offering a functional read-out for genetic variants: (6) provide a means to monitor response and recurrence of diseases, such as cancers: (7) describe the molecular landscape in human performance applications and extreme environments. Importantly, sophisticated metabolomic analytical platforms and informatics tools have recently been developed that make it possible to measure thousands of metabolites in blood, other body fluids, and tissues. Such tools also enable more robust analysis of response to treatment. New insights have been gained about mechanisms of diseases, including neuropsychiatric disorders, cardiovascular disease, cancers, diabetes and a range of pathologies. A series of ground breaking studies supported by National Institute of Health (NIH) through the Pharmacometabolomics Research Network and its partnership with the Pharmacogenomics Research Network illustrate how a patient's metabotype at baseline, prior to treatment, during treatment, and post-treatment, can inform about treatment outcomes and variations in responsiveness to drugs (e.g., statins, antidepressants, antihypertensives and antiplatelet therapies). These studies along with several others also exemplify how metabolomics data can complement and inform genetic data in defining ethnic, sex, and gender basis for variation in responses to treatment, which illustrates how pharmacometabolomics and pharmacogenomics are complementary and powerful tools for precision medicine.
Conclusions key scientific concepts and recommendations for precision medicine: Our metabolomics community believes that inclusion of metabolomics data in precision medicine initiatives is timely and will provide an extremely valuable layer of data that compliments and informs other data obtained by these important initiatives. Our Metabolomics Society, through its "Precision Medicine and Pharmacometabolomics Task Group", with input from our metabolomics community at large, has developed this White Paper where we discuss the value and approaches for including metabolomics data in large precision medicine initiatives. This White Paper offers recommendations for the selection of state of-the-art metabolomics platforms and approaches that offer the widest biochemical coverage, considers critical sample collection and preservation, as well as standardization of measurements, among other important topics. We anticipate that our metabolomics community will have representation in large precision medicine initiatives to provide input with regard to sample acquisition/preservation, selection of optimal omics technologies, and key issues regarding data collection, interpretation, and dissemination. We strongly recommend the collection and biobanking of samples for precision medicine initiatives that will take into consideration needs for large-scale metabolic phenotyping studies.
Keywords: Metabolomics; Metabonomics; Personalized medicine; Pharmacometabolomics; Pharmacometabonomics; Precision medicine.
Conflict of interest statement
The authors declare that they have no conflict of interest Ethical approval This is a review and does not contain any studies with human or animal subjects. Disclaimer for Dr. Beger The views expressed in this paper are solely those of the author, and they do not represent official policy of the U.S. Food and Drug Administration. Funding National Institute of General Medical Sciences (US); Award Numbers: R24 GM078233 and RC2GM092729 “The Pharmacometabolomics Research Network”; National Institute on Aging; Award numbers: 1R01AG046171; RF1AG051550 Grant Recipient: Rima Kaddurah-Daouk. In addition, this work was supported by funding to Phenome Centre Birmingham by the Medical Research Council in the UK (MR/M009157/1).
Figures
References
-
- Abo R, Hebbring S, Ji Y, Zhu H, Zeng ZB, Batzler A, et al. Merging pharmacometabolomics with pharmacogenomics using ‘1000 Genomes’ single-nucleotide polymorphism imputation: Selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenetics and Genomics. 2012;22(4):247–253. doi: 10.1097/FPC.0b013e32835001c9. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
