Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors
- PMID: 27643540
- PMCID: PMC6583788
- DOI: 10.1038/ng.3670
Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors
Abstract
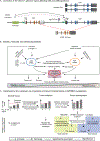
High rates of APOBEC-signature mutations are found in many tumors, but factors affecting this mutation pattern are not well understood. Here we explored the contribution of two common germline variants in the APOBEC3 region. SNP rs1014971 was associated with bladder cancer risk, increased APOBEC3B expression, and enrichment with APOBEC-signature mutations in bladder tumors. In contrast, a 30-kb deletion that eliminates APOBEC3B and creates an APOBEC3A-APOBEC3B chimera was not important in bladder cancer, whereas it was associated with breast cancer risk and enrichment with APOBEC-signature mutations in breast tumors. In vitro, APOBEC3B expression was predominantly induced by treatment with a DNA-damaging drug in bladder cancer cell lines, and APOBEC3A expression was induced as part of the antiviral interferon-stimulated response in breast cancer cell lines. These findings suggest a tissue-specific role of environmental oncogenic triggers, particularly in individuals with germline APOBEC3 risk variants.
Conflict of interest statement
Figures





Comment in
-
Re: Association of Germline Variants in the APOBEC3 Region with Cancer Risk and Enrichment with APOBEC-Signature Mutations in Tumors.J Urol. 2018 Jun;199(6):1394-1396. doi: 10.1016/j.juro.2018.03.016. Epub 2018 Mar 16. J Urol. 2018. PMID: 29783589 No abstract available.
References
Supplementary references:
-
- Marx V Drilling into big cancer-genome data. Nat Meth 10, 293–297 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

