Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein-protein crosslinks
- PMID: 27643814
- PMCID: PMC5192979
- DOI: 10.1002/pro.3045
Atomic modeling of the ITS2 ribosome assembly subcomplex from cryo-EM together with mass spectrometry-identified protein-protein crosslinks
Abstract
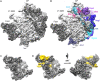
The assembly of ribosomal subunits starts in the nucleus, initiated by co-transcriptional folding of nascent ribosomal RNA (rRNA) transcripts and binding of ribosomal proteins and assembly factors. The internal transcribed spacer 2 (ITS2) is a precursor sequence to be processed from the intermediate 27S rRNA in the nucleoplasm; its removal is required for nuclear export of pre-60S particles. The proper processing of the ITS2 depends on multiple associated assembly factors and RNases. However, none of the structures of the known ITS2-binding factors is available. Here, we describe the modeling of the ITS2 subcomplex, including five assembly factors Cic1, Nop7, Nop15, Nop53, and Rlp7, using a combination of cryo-electron microscopy and cross-linking of proteins coupled with mass spectrometry approaches. The resulting atomic models provide structural insights into their function in ribosome assembly, and establish a framework for further dissection of their molecular roles in ITS2 processing.
Keywords: CXMS; Cic1; ITS2; Nop15; Nop53; Nop7; Rlp7; cross-linking; cryo-EM; ribosome assembly.
© 2016 The Protein Society.
Figures
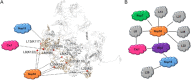


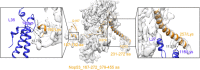


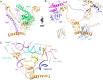
References
-
- Fernandez‐Pevida A, Kressler D, de la Cruz J (2015) Processing of preribosomal RNA in Saccharomyces cerevisiae . Wiley Interdiscip Rev RNA 6:191–209. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

