p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair
- PMID: 27644328
- PMCID: PMC5108885
- DOI: 10.1128/MCB.00434-16
p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair
Abstract
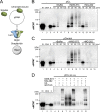
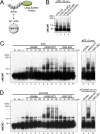
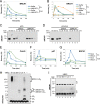
Interstrand cross-links (ICLs) are extremely toxic DNA lesions that create an impassable roadblock to DNA replication. When a replication fork collides with an ICL, it triggers a damage response that promotes multiple DNA processing events required to excise the cross-link from chromatin and resolve the stalled replication fork. One of the first steps in this process involves displacement of the CMG replicative helicase (comprised of Cdc45, MCM2-7, and GINS), which obstructs the underlying cross-link. Here we report that the p97/Cdc48/VCP segregase plays a critical role in ICL repair by unloading the CMG complex from chromatin. Eviction of the stalled helicase involves K48-linked polyubiquitylation of MCM7, p97-mediated extraction of CMG, and a largely degradation-independent mechanism of MCM7 deubiquitylation. Our results show that ICL repair and replication termination both utilize a similar mechanism to displace the CMG complex from chromatin. However, unlike termination, repair-mediated helicase unloading involves the tumor suppressor protein BRCA1, which acts upstream of MCM7 ubiquitylation and p97 recruitment. Together, these findings indicate that p97 plays a conserved role in dismantling the CMG helicase complex during different cellular events, but that distinct regulatory signals ultimately control when and where unloading takes place.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination.Genes Dev. 2017 Feb 1;31(3):275-290. doi: 10.1101/gad.291799.116. Epub 2017 Feb 24. Genes Dev. 2017. PMID: 28235849 Free PMC article.
-
Termination of DNA replication forks: "Breaking up is hard to do".Nucleus. 2015;6(3):187-96. doi: 10.1080/19491034.2015.1035843. Epub 2015 Apr 2. Nucleus. 2015. PMID: 25835602 Free PMC article.
-
Replication Fork Reversal during DNA Interstrand Crosslink Repair Requires CMG Unloading.Cell Rep. 2018 Jun 19;23(12):3419-3428. doi: 10.1016/j.celrep.2018.05.061. Cell Rep. 2018. PMID: 29924986 Free PMC article.
-
Should I stay or should I go: VCP/p97-mediated chromatin extraction in the DNA damage response.Exp Cell Res. 2014 Nov 15;329(1):9-17. doi: 10.1016/j.yexcr.2014.08.025. Epub 2014 Aug 27. Exp Cell Res. 2014. PMID: 25169698 Review.
-
Cdc48/p97 segregase: Spotlight on DNA-protein crosslinks.DNA Repair (Amst). 2024 Jul;139:103691. doi: 10.1016/j.dnarep.2024.103691. Epub 2024 May 9. DNA Repair (Amst). 2024. PMID: 38744091 Review.
Cited by
-
Coordinating DNA Replication and Mitosis through Ubiquitin/SUMO and CDK1.Int J Mol Sci. 2021 Aug 16;22(16):8796. doi: 10.3390/ijms22168796. Int J Mol Sci. 2021. PMID: 34445496 Free PMC article. Review.
-
Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.FEBS Lett. 2020 Mar;594(5):933-943. doi: 10.1002/1873-3468.13667. Epub 2019 Nov 22. FEBS Lett. 2020. PMID: 31701538 Free PMC article.
-
Proteolytic control of genome integrity at the replication fork.DNA Repair (Amst). 2019 Sep;81:102657. doi: 10.1016/j.dnarep.2019.102657. Epub 2019 Jul 10. DNA Repair (Amst). 2019. PMID: 31324531 Free PMC article. Review.
-
Fanconi anemia and the underlying causes of genomic instability.Environ Mol Mutagen. 2020 Aug;61(7):693-708. doi: 10.1002/em.22358. Epub 2020 Feb 6. Environ Mol Mutagen. 2020. PMID: 31983075 Free PMC article. Review.
-
The AAA+ ATPase p97, a cellular multitool.Biochem J. 2017 Aug 17;474(17):2953-2976. doi: 10.1042/BCJ20160783. Biochem J. 2017. PMID: 28819009 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
