Exploring the evolution of the proteins of the plant nuclear envelope
- PMID: 27644504
- PMCID: PMC5287204
- DOI: 10.1080/19491034.2016.1236166
Exploring the evolution of the proteins of the plant nuclear envelope
Abstract
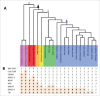
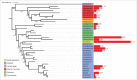
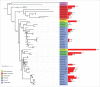
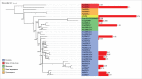
In this study, we explore the plasticity during evolution of proteins of the higher plant nuclear envelope (NE) from the most ancestral plant species to advanced angiosperms. The higher plant NE contains a functional Linker of Nucleoskeleton and Cytoskeleton (LINC) complex based on conserved Sad1-Unc84 (SUN) domain proteins and plant specific Klarsicht/Anc1/Syne homology (KASH) domain proteins. Recent evidence suggests the presence of a plant lamina underneath the inner membrane and various coiled-coil proteins have been hypothesized to be associated with it including Crowded Nuclei (CRWN; also termed LINC and NMCP), Nuclear Envelope Associated Protein (NEAP) protein families as well as the CRWN binding protein KAKU4. SUN domain proteins appear throughout with a key role for mid-SUN proteins suggested. Evolution of KASH domain proteins has resulted in increasing complexity, with some appearing in all species considered, while other KASH proteins are progressively gained during evolution. Failure to identify CRWN homologs in unicellular organisms included in the study and their presence in plants leads us to speculate that convergent evolution may have occurred in the formation of the lamina with each kingdom having new proteins such as the Lamin B receptor (LBR) and Lamin-Emerin-Man1 (LEM) domain proteins (animals) or NEAPs and KAKU4 (plants). Our data support a model in which increasing complexity at the nuclear envelope occurred through the plant lineage and suggest a key role for mid-SUN proteins as an early and essential component of the nuclear envelope.
Keywords: Chromatin; KASH domain; LINC complex; SUN domain; higher plant; nucleoskeleton; nucleus.
Figures

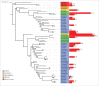
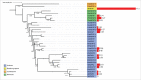
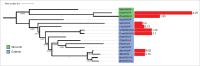
References
-
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al.. The revised classification of eukaryotes. J Eukaryot Microbiol 2012; 59:429-514; PMID:23020233; http://dx.doi.org/10.1111/j.1550-7408.2012.00644.x - DOI - PMC - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-410; PMID:2231712; http://dx.doi.org/10.1016/S0022-2836(05)80360-2 - DOI - PubMed
-
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166-9; PMID:25260700; http://dx.doi.org/10.1093/bioinformatics/btu638 - DOI - PMC - PubMed
-
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 2013; 152:1270-84; PMID:23498936 - PubMed
-
- Cavalier-Smith T. Kingdoms protozoa and chromista and the eozoan root of the eukaryotic tree. Biol Lett 2010; 6:342-5; PMID:20031978; http://dx.doi.org/10.1098/rsbl.2009.0948 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
