Identification of 2-oxohistidine Interacting Proteins Using E. coli Proteome Chips
- PMID: 27644758
- PMCID: PMC5141273
- DOI: 10.1074/mcp.M116.060806
Identification of 2-oxohistidine Interacting Proteins Using E. coli Proteome Chips
Abstract
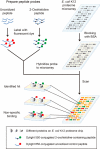
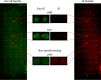
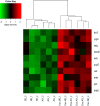
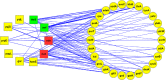
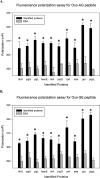
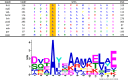
Cellular proteins are constantly damaged by reactive oxygen species generated by cellular respiration. Because of its metal-chelating property, the histidine residue is easily oxidized in the presence of Cu/Fe ions and H2O2 via metal-catalyzed oxidation, usually converted to 2-oxohistidine. We hypothesized that cells may have evolved antioxidant defenses against the generation of 2-oxohistidine residues on proteins, and therefore there would be cellular proteins which specifically interact with this oxidized side chain. Using two chemically synthesized peptide probes containing 2-oxohistidine, high-throughput interactome screening was conducted using the E. coli K12 proteome microarray containing >4200 proteins. Ten interacting proteins were identified, and successfully validated using a third peptide probe, fluorescence polarization assays, as well as binding constant measurements. We discovered that 9 out of 10 identified proteins seemed to be involved in redox-related cellular functions. We also built the functional interaction network to reveal their interacting proteins. The network showed that our interacting proteins were enriched in oxido-reduction processes, ion binding, and carbon metabolism. A consensus motif was identified among these 10 bacterial interacting proteins based on bioinformatic analysis, which also appeared to be present on human S100A1 protein. Besides, we found that the consensus binding motif among our identified proteins, including bacteria and human, were located within α-helices and faced the outside of proteins. The combination of chemically engineered peptide probes with proteome microarrays proves to be an efficient discovery platform for protein interactomes of unusual post-translational modifications, and sensitive enough to detect even the insertion of a single oxygen atom in this case.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Wold F. (1981) In vivo chemical modification of proteins (post-translational modification). Annu. Rev. Biochem. 50, 783–814 - PubMed
-
- Walsh C. T., Garneau-Tsodikova S., and Gatto G. J. Jr. (2005) Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed. 44, 7342–7372 - PubMed
-
- Harding J. J. (1985) Nonenzymatic covalent posttranslational modification of proteins in vivo. Adv. Protein Chem. 37, 247–334 - PubMed
-
- Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

