Quantitative proteomics and terminomics to elucidate the role of ubiquitination and proteolysis in adaptive immunity
- PMID: 27644975
- PMCID: PMC5031638
- DOI: 10.1098/rsta.2015.0372
Quantitative proteomics and terminomics to elucidate the role of ubiquitination and proteolysis in adaptive immunity
Abstract
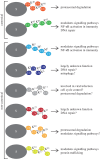
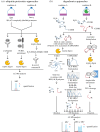
Adaptive immunity is the specialized defence mechanism in vertebrates that evolved to eliminate pathogens. Specialized lymphocytes recognize specific protein epitopes through antigen receptors to mount potent immune responses, many of which are initiated by nuclear factor-kappa B activation and gene transcription. Most, if not all, pathways in adaptive immunity are further regulated by post-translational modification (PTM) of signalling proteins, e.g. phosphorylation, citrullination, ubiquitination and proteolytic processing. The importance of PTMs is reflected by genetic or acquired defects in these pathways that lead to a dysfunctional immune response. Here we discuss the state of the art in targeted proteomics and systems biology approaches to dissect the PTM landscape specifically regarding ubiquitination and proteolysis in B- and T-cell activation. Recent advances have occurred in methods for specific enrichment and targeted quantitation. Together with improved instrument sensitivity, these advances enable the accurate analysis of often rare PTM events that are opaque to conventional proteomics approaches, now rendering in-depth analysis and pathway dissection possible. We discuss published approaches, including as a case study the profiling of the N-terminome of lymphocytes of a rare patient with a genetic defect in the paracaspase protease MALT1, a key regulator protease in antigen-driven signalling, which was manifested by elevated linear ubiquitination.This article is part of the themed issue 'Quantitative mass spectrometry'.
Keywords: LUBAC; MALT1; degradomics; linear ubiquitin; proteases; ubiquitin.
© 2016 The Authors.
Figures
Similar articles
-
The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF-κB signalling.Nat Commun. 2015 Nov 3;6:8777. doi: 10.1038/ncomms9777. Nat Commun. 2015. PMID: 26525107 Free PMC article.
-
Combinatorial degradomics: Precision tools to unveil proteolytic processes in biological systems.Biochim Biophys Acta Proteins Proteom. 2020 Jun;1868(6):140392. doi: 10.1016/j.bbapap.2020.140392. Epub 2020 Feb 20. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32087360 Review.
-
Characterization of ubiquitination dependent dynamics in growth factor receptor signaling by quantitative proteomics.Mol Biosyst. 2011 Dec;7(12):3223-33. doi: 10.1039/c1mb05185g. Epub 2011 Sep 28. Mol Biosyst. 2011. PMID: 21956701
-
Top-down proteomics for the analysis of proteolytic events - Methods, applications and perspectives.Biochim Biophys Acta Mol Cell Res. 2017 Nov;1864(11 Pt B):2191-2199. doi: 10.1016/j.bbamcr.2017.07.002. Epub 2017 Jul 12. Biochim Biophys Acta Mol Cell Res. 2017. PMID: 28711385 Review.
-
N- and C-terminal degradomics: new approaches to reveal biological roles for plant proteases from substrate identification.Physiol Plant. 2012 May;145(1):5-17. doi: 10.1111/j.1399-3054.2011.01536.x. Epub 2011 Dec 7. Physiol Plant. 2012. PMID: 22023699 Review.
Cited by
-
Proteomic analysis of the cardiac extracellular matrix: clinical research applications.Expert Rev Proteomics. 2018 Feb;15(2):105-112. doi: 10.1080/14789450.2018.1421947. Epub 2018 Jan 9. Expert Rev Proteomics. 2018. PMID: 29285949 Free PMC article. Review.
-
Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia.Sci Rep. 2019 Feb 19;9(1):2307. doi: 10.1038/s41598-019-38490-1. Sci Rep. 2019. PMID: 30783160 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

