Oxford Nanopore MinION Sequencing and Genome Assembly
- PMID: 27646134
- PMCID: PMC5093776
- DOI: 10.1016/j.gpb.2016.05.004
Oxford Nanopore MinION Sequencing and Genome Assembly
Abstract
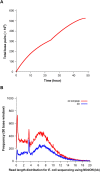
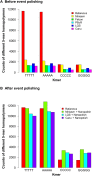
The revolution of genome sequencing is continuing after the successful second-generation sequencing (SGS) technology. The third-generation sequencing (TGS) technology, led by Pacific Biosciences (PacBio), is progressing rapidly, moving from a technology once only capable of providing data for small genome analysis, or for performing targeted screening, to one that promises high quality de novo assembly and structural variation detection for human-sized genomes. In 2014, the MinION, the first commercial sequencer using nanopore technology, was released by Oxford Nanopore Technologies (ONT). MinION identifies DNA bases by measuring the changes in electrical conductivity generated as DNA strands pass through a biological pore. Its portability, affordability, and speed in data production makes it suitable for real-time applications, the release of the long read sequencer MinION has thus generated much excitement and interest in the genomics community. While de novo genome assemblies can be cheaply produced from SGS data, assembly continuity is often relatively poor, due to the limited ability of short reads to handle long repeats. Assembly quality can be greatly improved by using TGS long reads, since repetitive regions can be easily expanded into using longer sequencing lengths, despite having higher error rates at the base level. The potential of nanopore sequencing has been demonstrated by various studies in genome surveillance at locations where rapid and reliable sequencing is needed, but where resources are limited.
Keywords: De novo assembly; Molecular clinical diagnostics; Oxford nanopore MinION device; Structural variations; Third-generation sequencing.
Copyright © 2016 The Authors. Production and hosting by Elsevier Ltd.. All rights reserved.
Figures


References
-
- The International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Sanger F., Coulson A.R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

