Origin of the plant Tm-1-like gene via two independent horizontal transfer events and one gene fusion event
- PMID: 27647002
- PMCID: PMC5028733
- DOI: 10.1038/srep33691
Origin of the plant Tm-1-like gene via two independent horizontal transfer events and one gene fusion event
Abstract
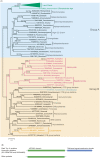
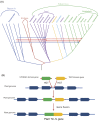
The Tomato mosaic virus (ToMV) resistance gene Tm-1 encodes a direct inhibitor of ToMV RNA replication to protect tomato from infection. The plant Tm-1-like (Tm-1L) protein is predicted to contain an uncharacterized N-terminal UPF0261 domain and a C-terminal TIM-barrel signal transduction (TBST) domain. Homologous searches revealed that proteins containing both of these two domains are mainly present in charophyte green algae and land plants but absent from glaucophytes, red algae and chlorophyte green algae. Although Tm-1 homologs are widely present in bacteria, archaea and fungi, UPF0261- and TBST-domain-containing proteins are generally encoded by different genes in these linages. A co-evolution analysis also suggested a putative interaction between UPF0261- and TBST-domain-containing proteins. Phylogenetic analyses based on homologs of these two domains revealed that plants have acquired UPF0261- and TBST-domain-encoding genes through two independent horizontal gene transfer (HGT) events before the origin of land plants from charophytes. Subsequently, gene fusion occurred between these two horizontally acquired genes and resulted in the origin of the Tm-1L gene in streptophytes. Our results demonstrate a novel evolutionary mechanism through which the recipient organism may acquire genes with functional interaction through two different HGT events and further fuse them into one functional gene.
Figures


Similar articles
-
Coevolution and hierarchical interactions of Tomato mosaic virus and the resistance gene Tm-1.PLoS Pathog. 2012;8(10):e1002975. doi: 10.1371/journal.ppat.1002975. Epub 2012 Oct 18. PLoS Pathog. 2012. PMID: 23093939 Free PMC article.
-
An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses.Proc Natl Acad Sci U S A. 2009 May 26;106(21):8778-83. doi: 10.1073/pnas.0809105106. Epub 2009 May 7. Proc Natl Acad Sci U S A. 2009. PMID: 19423673 Free PMC article.
-
An inhibitor of viral RNA replication is encoded by a plant resistance gene.Proc Natl Acad Sci U S A. 2007 Aug 21;104(34):13833-8. doi: 10.1073/pnas.0703203104. Epub 2007 Aug 15. Proc Natl Acad Sci U S A. 2007. PMID: 17699618 Free PMC article.
-
Mechanisms of tomato mosaic virus RNA replication and its inhibition by the host resistance factor Tm-1.Curr Opin Virol. 2014 Dec;9:8-13. doi: 10.1016/j.coviro.2014.08.005. Epub 2014 Sep 17. Curr Opin Virol. 2014. PMID: 25212767 Review.
-
Adaptive innovation of green plants by horizontal gene transfer.Biotechnol Adv. 2021 Jan-Feb;46:107671. doi: 10.1016/j.biotechadv.2020.107671. Epub 2020 Nov 24. Biotechnol Adv. 2021. PMID: 33242576 Review.
Cited by
-
Potential Effects of Horizontal Gene Exchange in the Human Gut.Front Immunol. 2017 Nov 27;8:1630. doi: 10.3389/fimmu.2017.01630. eCollection 2017. Front Immunol. 2017. PMID: 29230215 Free PMC article.
-
Ancestor of land plants acquired the DNA-3-methyladenine glycosylase (MAG) gene from bacteria through horizontal gene transfer.Sci Rep. 2017 Aug 24;7(1):9324. doi: 10.1038/s41598-017-05066-w. Sci Rep. 2017. PMID: 28839126 Free PMC article.
-
Horizontal Gene Transfer and Fusion Spread Carotenogenesis Among Diverse Heterotrophic Protists.Genome Biol Evol. 2023 Mar 3;15(3):evad029. doi: 10.1093/gbe/evad029. Genome Biol Evol. 2023. PMID: 36805209 Free PMC article.
-
Immune Receptors and Co-receptors in Antiviral Innate Immunity in Plants.Front Microbiol. 2017 Jan 5;7:2139. doi: 10.3389/fmicb.2016.02139. eCollection 2016. Front Microbiol. 2017. PMID: 28105028 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

