Dpy30 is critical for maintaining the identity and function of adult hematopoietic stem cells
- PMID: 27647347
- PMCID: PMC5068233
- DOI: 10.1084/jem.20160185
Dpy30 is critical for maintaining the identity and function of adult hematopoietic stem cells
Abstract
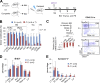
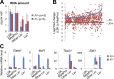
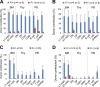
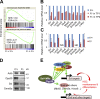
As the major histone H3K4 methyltransferases in mammals, the Set1/Mll complexes play important roles in animal development and are associated with many diseases, including hematological malignancies. However, the role of the H3K4 methylation activity of these complexes in fate determination of hematopoietic stem and progenitor cells (HSCs and HPCs) remains elusive. Here, we address this question by generating a conditional knockout mouse for Dpy30, which is a common core subunit of all Set1/Mll complexes and facilitates genome-wide H3K4 methylation in cells. Dpy30 loss in the adult hematopoietic system results in severe pancytopenia but striking accumulation of HSCs and early HPCs that are defective in multilineage reconstitution, suggesting a differentiation block. In mixed bone marrow chimeras, Dpy30-deficient HSCs cannot differentiate or efficiently up-regulate lineage-regulatory genes, and eventually fail to sustain for long term with significant loss of HSC signature gene expression. Our molecular analyses reveal that Dpy30 directly and preferentially controls H3K4 methylation and expression of many hematopoietic development-associated genes including several key transcriptional and chromatin regulators involved in HSC function. Collectively, our results establish a critical and selective role of Dpy30 and the H3K4 methylation activity of the Set1/Mll complexes for maintaining the identity and function of adult HSCs.
© 2016 Yang et al.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

