Dual T cell- and B cell-intrinsic deficiency in humans with biallelic RLTPR mutations
- PMID: 27647349
- PMCID: PMC5068239
- DOI: 10.1084/jem.20160576
Dual T cell- and B cell-intrinsic deficiency in humans with biallelic RLTPR mutations
Abstract
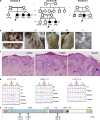
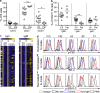
Combined immunodeficiency (CID) refers to inborn errors of human T cells that also affect B cells because of the T cell deficit or an additional B cell-intrinsic deficit. In this study, we report six patients from three unrelated families with biallelic loss-of-function mutations in RLTPR, the mouse orthologue of which is essential for CD28 signaling. The patients have cutaneous and pulmonary allergy, as well as a variety of bacterial and fungal infectious diseases, including invasive tuberculosis and mucocutaneous candidiasis. Proportions of circulating regulatory T cells and memory CD4+ T cells are reduced. Their CD4+ T cells do not respond to CD28 stimulation. Their CD4+ T cells exhibit a "Th2" cell bias ex vivo and when cultured in vitro, contrasting with the paucity of "Th1," "Th17," and T follicular helper cells. The patients also display few memory B cells and poor antibody responses. This B cell phenotype does not result solely from the T cell deficiency, as the patients' B cells fail to activate NF-κB upon B cell receptor (BCR) stimulation. Human RLTPR deficiency is a CID affecting at least the CD28-responsive pathway in T cells and the BCR-responsive pathway in B cells.
© 2016 Wang et al.
Figures





References
-
- Alkhairy O.K., Perez-Becker R., Driessen G.J., Abolhassani H., van Montfrans J., Borte S., Choo S., Wang N., Tesselaar K., Fang M., et al. 2015. Novel mutations in TNFRSF7/CD27: Clinical, immunologic, and genetic characterization of human CD27 deficiency. J. Allergy Clin. Immunol. 136:703–712.e10. 10.1016/j.jaci.2015.02.022 - DOI - PubMed
-
- Altin J.A., Tian L., Liston A., Bertram E.M., Goodnow C.C., and Cook M.C.. 2011. Decreased T-cell receptor signaling through Card11 differentially compromises forkhead box protein 3–positive regulatory versus TH2 effector cells to cause allergy. J. Allergy Clin. Immunol. 127:1277–1285.e5. 10.1016/j.jaci.2010.12.1081 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

