Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization
- PMID: 27647905
- PMCID: PMC5056070
- DOI: 10.1073/pnas.1607855113
Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization
Abstract
Defensins are cationic antimicrobial peptides that serve as important components of host innate immune defenses, often by targeting cell membranes of pathogens. Oligomerization of defensins has been linked to their antimicrobial activity; however, the molecular basis underpinning this process remains largely unclear. Here we show that the plant defensin NsD7 targets the phospholipid phosphatidic acid (PA) to form oligomeric complexes that permeabilize PA-containing membranes. The crystal structure of the NsD7-PA complex reveals a striking double helix of two right-handed coiled oligomeric defensin fibrils, the assembly of which is dependent upon the interaction with PA at the interface between NsD7 dimers. Using site-directed mutagenesis, we demonstrate that key residues in this PA-binding site are required for PA-mediated NsD7 oligomerization and coil formation, as well as permeabilization of PA-containing liposomes. These data suggest that multiple lipids can be targeted to induce oligomerization of defensins during membrane permeabilization and demonstrate the existence of a "phospholipid code" that identifies target membranes for defensin-mediated attack as part of a first line of defense across multiple species.
Keywords: antimicrobial peptides; defensins; host–pathogen interactions; innate defense; phospholipids.
Conflict of interest statement
M.D.H. is a former Vice President of Research in Hexima Ltd.
Figures
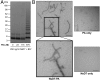
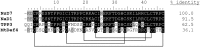
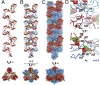
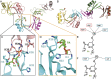


References
-
- Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. - PubMed
-
- Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem. 1999;266(1):1–16. - PubMed
-
- Testerink C, Munnik T. Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10(8):368–375. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

