Whole-exome sequencing identifies multiple loss-of-function mutations of NF-κB pathway regulators in nasopharyngeal carcinoma
- PMID: 27647909
- PMCID: PMC5056105
- DOI: 10.1073/pnas.1607606113
Whole-exome sequencing identifies multiple loss-of-function mutations of NF-κB pathway regulators in nasopharyngeal carcinoma
Abstract
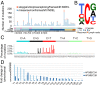
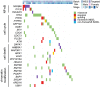
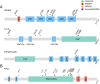
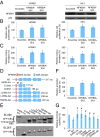
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy with a unique geographical distribution. The genomic abnormalities leading to NPC pathogenesis remain unclear. In total, 135 NPC tumors were examined to characterize the mutational landscape using whole-exome sequencing and targeted resequencing. An APOBEC cytidine deaminase mutagenesis signature was revealed in the somatic mutations. Noticeably, multiple loss-of-function mutations were identified in several NF-κB signaling negative regulators NFKBIA, CYLD, and TNFAIP3 Functional studies confirmed that inhibition of NFKBIA had a significant impact on NF-κB activity and NPC cell growth. The identified loss-of-function mutations in NFKBIA leading to protein truncation contributed to the altered NF-κB activity, which is critical for NPC tumorigenesis. In addition, somatic mutations were found in several cancer-relevant pathways, including cell cycle-phase transition, cell death, EBV infection, and viral carcinogenesis. These data provide an enhanced road map for understanding the molecular basis underlying NPC.
Keywords: APOBEC-mediated signature; NF-κB signaling; nasopharyngeal carcinoma; somatic mutation landscape; whole-exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bruce JP, Yip K, Bratman SV, Ito E, Liu FF. Nasopharyngeal cancer: Molecular landscape. J Clin Oncol. 2015;33(29):3346–3355. - PubMed
-
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol. 2012;22(2):79–86. - PubMed
-
- Lin DC, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

