Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition
- PMID: 27647925
- PMCID: PMC5056096
- DOI: 10.1073/pnas.1612579113
Cell cycle progression in Caulobacter requires a nucleoid-associated protein with high AT sequence recognition
Abstract
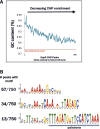
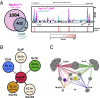
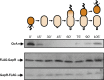
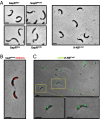
Faithful cell cycle progression in the dimorphic bacterium Caulobacter crescentus requires spatiotemporal regulation of gene expression and cell pole differentiation. We discovered an essential DNA-associated protein, GapR, that is required for Caulobacter growth and asymmetric division. GapR interacts with adenine and thymine (AT)-rich chromosomal loci, associates with the promoter regions of cell cycle-regulated genes, and shares hundreds of recognition sites in common with known master regulators of cell cycle-dependent gene expression. GapR target loci are especially enriched in binding sites for the transcription factors GcrA and CtrA and overlap with nearly all of the binding sites for MucR1, a regulator that controls the establishment of swarmer cell fate. Despite constitutive synthesis, GapR accumulates preferentially in the swarmer compartment of the predivisional cell. Homologs of GapR, which are ubiquitous among the α-proteobacteria and are encoded on multiple bacteriophage genomes, also accumulate in the predivisional cell swarmer compartment when expressed in Caulobacter The Escherichia coli nucleoid-associated protein H-NS, like GapR, selectively associates with AT-rich DNA, yet it does not localize preferentially to the swarmer compartment when expressed exogenously in Caulobacter, suggesting that recognition of AT-rich DNA is not sufficient for the asymmetric accumulation of GapR. Further, GapR does not silence the expression of H-NS target genes when expressed in E. coli, suggesting that GapR and H-NS have distinct functions. We propose that Caulobacter has co-opted a nucleoid-associated protein with high AT recognition to serve as a mediator of cell cycle progression.
Keywords: AT-rich; Caulobacter; asymmetry; cell cycle; nucleoid-associated protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
Replication fork passage drives asymmetric dynamics of a critical nucleoid-associated protein in Caulobacter.EMBO J. 2017 Feb 1;36(3):301-318. doi: 10.15252/embj.201695513. Epub 2016 Dec 23. EMBO J. 2017. PMID: 28011580 Free PMC article.
-
Regulation of the activity of the dual-function DnaA protein in Caulobacter crescentus.PLoS One. 2011;6(10):e26028. doi: 10.1371/journal.pone.0026028. Epub 2011 Oct 14. PLoS One. 2011. PMID: 22022497 Free PMC article.
-
A novel nucleoid-associated protein coordinates chromosome replication and chromosome partition.Nucleic Acids Res. 2017 Sep 6;45(15):8916-8929. doi: 10.1093/nar/gkx596. Nucleic Acids Res. 2017. PMID: 28911105 Free PMC article.
-
Regulation of chromosomal replication in Caulobacter crescentus.Plasmid. 2012 Mar;67(2):76-87. doi: 10.1016/j.plasmid.2011.12.007. Epub 2011 Dec 29. Plasmid. 2012. PMID: 22227374 Review.
-
Cell cycle regulation in Caulobacter: location, location, location.J Cell Sci. 2007 Oct 15;120(Pt 20):3501-7. doi: 10.1242/jcs.005967. J Cell Sci. 2007. PMID: 17928306 Review.
Cited by
-
Molecular architecture of the DNA-binding sites of the P-loop ATPases MipZ and ParA from Caulobacter crescentus.Nucleic Acids Res. 2020 May 21;48(9):4769-4779. doi: 10.1093/nar/gkaa192. Nucleic Acids Res. 2020. PMID: 32232335 Free PMC article.
-
The Caulobacter NtrB-NtrC two-component system bridges nitrogen assimilation and cell development.J Bacteriol. 2023 Oct 26;205(10):e0018123. doi: 10.1128/jb.00181-23. Epub 2023 Oct 4. J Bacteriol. 2023. PMID: 37791753 Free PMC article.
-
DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity.DNA (Basel). 2024 Nov 19;4(4):473-493. doi: 10.3390/dna4040032. DNA (Basel). 2024. PMID: 40098770 Free PMC article.
-
Compaction and control-the role of chromosome-organizing proteins in Streptomyces.FEMS Microbiol Rev. 2020 Nov 24;44(6):725-739. doi: 10.1093/femsre/fuaa028. FEMS Microbiol Rev. 2020. PMID: 32658291 Free PMC article. Review.
-
LdtR is a master regulator of gene expression in Liberibacter asiaticus.Microb Biotechnol. 2017 Jul;10(4):896-909. doi: 10.1111/1751-7915.12728. Epub 2017 May 15. Microb Biotechnol. 2017. PMID: 28503858 Free PMC article.
References
-
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8(3):185–195. - PubMed
-
- Dorman CJ. Genome architecture and global gene regulation in bacteria: Making progress towards a unified model? Nat Rev Microbiol. 2013;11(5):349–355. - PubMed
-
- Marbouty M, et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol Cell. 2015;59(4):588–602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

